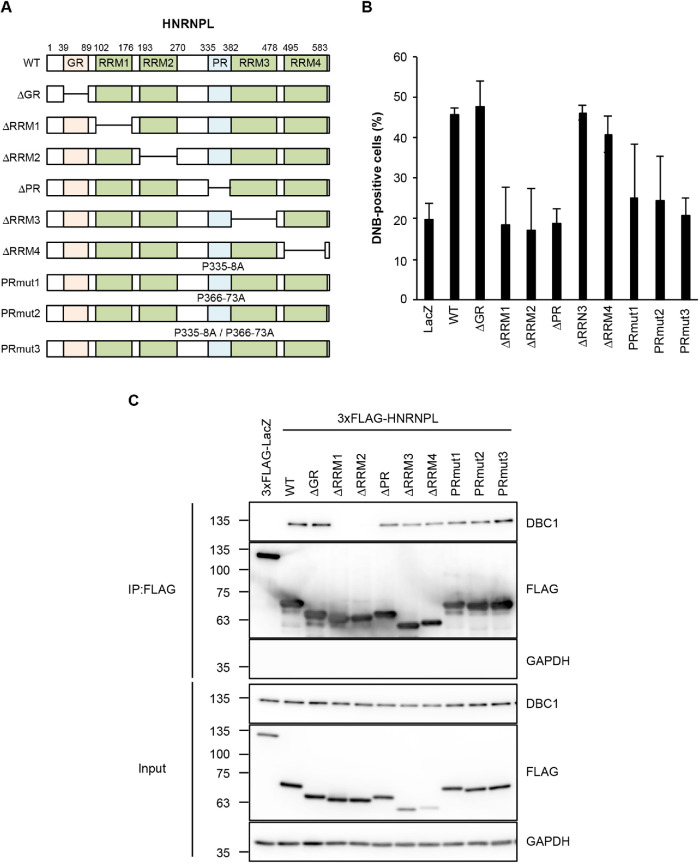
FIGURE 4:
The RNA-binding domain and PR of HNRNPL are required for DNB formation. (A) Schematic presentation of both deletion mutants and point mutants of HNRNPL. For three point mutants (PRmut1, PRmut2, and PRmut3), the mutated residues are shown. (B) RRM1, RRM2, and PR are required for DNB formation. Rescue of the defect in DNB formation by the HNRNPL mutant constructs shown in A. The HNRNPL constructs were transfected into HCT116 cells in which the endogenous HNRNPL has been depleted by RNAi, and then DNB-positive cells (DBC1 foci–positive cells) were counted (>100 cells, ±SD, n = 5). As a negative control, the FLAG-LacZ plasmid was transfected (LacZ). (C) Identification of the HNRNPL domains required for the interaction with DBC1. A series of FLAG-tagged HNRNPL mutants were immunoprecipitated, and coprecipitated DBC1 was detected by Western blotting. GAPDH denotes the input control. The molecular mass marker (kDa) is shown on the left.