Abstract
Copper homeostasis is crucial for cellular physiology and development, and its dysregulation leads to disease. The Menkes ATPase ATP7A plays a key role in copper efflux, by trafficking from the Golgi to the plasma membrane upon cell exposure to elevated copper, but the mechanisms that target ATP7A to the cell periphery are poorly understood. PDZD11 interacts with the C-terminus of ATP7A, which contains sequences involved in ATP7A trafficking, but the role of PDZD11 in ATP7A localization is unknown. Here we identify PLEKHA5 and PLEKHA6 as new interactors of PDZD11 that bind to the PDZD11 N-terminus through their WW domains similarly to the junctional protein PLEKHA7. Using CRISPR-KO kidney epithelial cells, we show by immunofluorescence microscopy that WW-PLEKHAs (PLEKHA5, PLEKHA6, PLEKHA7) recruit PDZD11 to distinct plasma membrane localizations and that they are required for the efficient anterograde targeting of ATP7A to the cell periphery in elevated copper conditions. Pull-down experiments show that WW-PLEKHAs promote PDZD11 interaction with the C-terminus of ATP7A. However, WW-PLEKHAs and PDZD11 are not necessary for ATP7A Golgi localization in basal copper, ATP7A copper-induced exit from the Golgi, and ATP7A retrograde trafficking to the Golgi. Finally, measuring bioavailable and total cellular copper, metallothionein-1 expression, and cell viability shows that WW-PLEKHAs and PDZD11 are required for maintaining low intracellular copper levels when cells are exposed to elevated copper. These data indicate that WW-PLEKHAs-PDZD11 complexes regulate the localization and function of ATP7A to promote copper extrusion in elevated copper.
INTRODUCTION
Copper is an essential micronutrient as a cofactor of vital oxidative enzymes, and both its deficiency and overload are pathological, as exemplified by the Menkes and Wilson diseases (Bandmann et al., 2015; Czlonkowska et al., 2018). The brain and nervous tissue have a particularly high oxidative demand and requirement for copper-dependent enzyme activities, and several neurological disorders, including Alzheimer’s, Parkinson’s, and Huntington’s diseases, are associated with disrupted copper homeostasis (Kaler, 2011; Telianidis et al., 2013; Zlatic et al., 2015; Ackerman and Chang, 2018; Lutsenko et al., 2019). Intracellular copper is physiologically finely tuned through the coordination of copper uptake, intracellular trafficking by copper chaperones, and efflux by specific copper efflux transporters (reviewed in Camakaris et al., 1999; La Fontaine and Mercer, 2007; Lutsenko et al., 2007; Polishchuk and Lutsenko, 2013; Hartwig et al., 2019). The P-type ATPases ATP7A (Menkes copper ATPase) and ATP7B (Wilson copper ATPase) play a major role in copper efflux in different tissues (La Fontaine and Mercer, 2007; Lutsenko et al., 2007; Nevitt et al., 2012). Under basal copper conditions, ATP7A is localized in a specific compartment of the trans-Golgi network (TGN), whereas elevated copper levels induce its translocation from the TGN to a compartment of post-Golgi membrane vesicles that are trafficked to the plasma membrane and whose exocytosis and endocytic recycling drive copper efflux (Camakaris et al., 1995; Petris et al., 1996; 2002; Petris and Mercer, 1999; Yamaguchi et al., 1996; Dierick et al., 1997; La Fontaine et al., 1998, 1999; Cobbold et al., 2002; Monty et al., 2005; Holloway et al., 2007; Nyasae et al., 2007).
Although the severity of Menkes disease correlates with the intracellular localization of ATP7A (Skjorringe et al., 2017), the molecular mechanisms that regulate the trafficking and plasma membrane localization of ATP7A-containing vesicles remain unresolved. In cultured polarized epithelial cells, C-terminal di-leucine and PDZ-binding motifs are required for basolateral plasma membrane targeting of ATP7A in elevated copper (Greenough et al., 2004). The single PDZ domain-containing protein 11 (PDZD11, also known as AIPP1) was identified as an interactor of the C-terminal 15 amino acids of ATP7A, and in Saccharomyces cerevisiae this interaction was not dependent on copper levels (Stephenson et al., 2005). Although these observations suggest that PDZD11 could be involved in plasma membrane targeting of ATP7A, the role of PDZD11 in ATP7A trafficking and in copper homeostasis has not been explored.
We identified PDZD11 as an interactor of the adherens junction (AJ) protein PLEKHA7 (Guerrera et al., 2016). The N-terminal tandem WW domains of PLEKHA7 bind to the N-terminal proline-rich sequence of PDZD11 to recruit PDZD11 to the AJ, and this interaction is required for the accumulation of the transmembrane proteins nectins, tetraspanin-33 (Tspan33), and ADAM10 at the AJ (Guerrera et al., 2016; Shah et al., 2018; Rouaud et al., 2020). Although we detected PDZD11 labeling predominantly at cell–cell junctions using antibodies against the endogenous protein (Guerrera et al., 2016), PDZD11 was reported to interact not only with ATP7A (Stephenson et al., 2005), but also with additional transmembrane transporters, such as the plasma membrane calcium ATPase (PMCA) and the sodium-dependent multivitamin transporter SLC5A6 (Goellner et al., 2003; Nabokina et al., 2011), which are localized not at junctions but along the basolateral membrane of polarized epithelial cells (Chicka and Strehler, 2003; Greenough et al., 2004; Subramanian et al., 2009). This suggests that PDZD11 interacts with other ligands at these sites independently of PLEKHA7. To address this question and gain more insight into the molecular complexes that implicate PDZD11, we searched for new PDZD11 interactors through a yeast two-hybrid (Y2H) screen. We report here that PLEKHA5 (Pleckstrin homology domain–containing family A member 5) and PLEKHA6 (Pleckstrin homology domain–containing family A member 6), also known respectively as phosphatidylinositol-three-phosphate-binding PH-domain protein-2 (PEPP2) and -3 (PEPP3) (Dowler et al., 2000), are new PDZD11-interacting proteins that are localized in the cytoplasm and near the apical, lateral, and basal plasma membranes in epithelial cells within tissues and in culture. Because PDZD11 binds to ATP7A sequences required for its trafficking to the cell periphery (Greenough et al., 2004; Stephenson et al., 2005), we hypothesized that PDZD11, PLEKHA5, PLEKHA6, and PLEKHA7 are involved in ATP7A trafficking. Here we validate this hypothesis, and we show that PLEKHA5, PLEKHA6, and PLEKHA7 promote the binding of PDZD11 to the C-terminus of ATP7A and together with PDZD11 are also required for efficient copper efflux upon cell exposure to elevated copper.
RESULTS
PLEKHA5 and PLEKHA6 are new interactors of PDZD11
In addition to interactions with known partners, that is, PLEKHA7 (residues 1–113 of transcript variant NM_175058.4) (Guerrera et al., 2016) and the sodium-dependent multivitamin transporter SLC5A6 (Nabokina et al., 2011), a Y2H screen using full-length PDZD11 as a bait for a human placenta library containing fragments of cDNAs revealed high confidence interactions with the N-terminal sequences of PLEKHA5 (residues 1–117 of transcript variant 1, NM_019012.5) and PLEKHA6 (residues 1–93 of transcript variant X11, XM_011509297.2) (Figure 1A). Sequence analysis shows that similarly to PLEKHA7, and unlike other members of the PLEKHA family of proteins, these PLEKHA5 and PLEKHA6 isoforms contain N-terminal tandem WW domains. The highest degree of sequence similarity is displayed by the N-terminal regions, comprising the WW and PH domains (Supplemental Figure S1, A–E). We propose the name WW-PLEKHAs for these members of the PLEKHA family that contain tandem WW domains. WW-PLEKHAs also comprise a PH domain, typical of the PLEKHA family, and C-terminal regions that contain coiled-coil (CC) and proline-rich (Pro-rich) domains (Figure 1A; Supplemental Figure S1A).
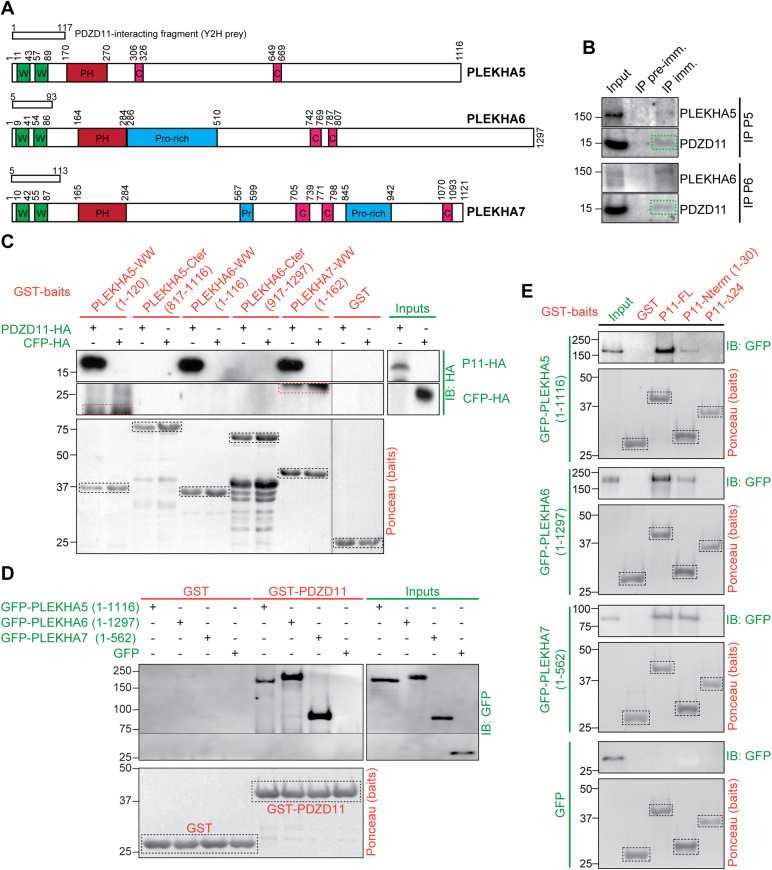
FIGURE 1:
PLEKHA5 and PLEKHA6 interact with the N-terminus of PDZD11 through their tandem WW domains. (A) Schemes of human PLEKHA5, PLEKHA6, and PLEKHA7 showing amino acid positions and structural domains: WW (Trp-Trp) green, PH (pleckstrin homology) red, proline-rich (Pro-rich) blue, coiled-coil (CC) pink. Boxes show yeast two-hybrid (Y2H) screen preys. (B) Representative immunoblot (IB) analysis of PDZD11 in immunoprecipitates (IP) of either PLEKHA5 (P5) or PLEKHA6 (P6) from lysates of Caco-2 cells (preimmune serum as negative control) (n = 4 for P5, n = 3 for P6). (C) IB analysis, using anti-HA antibody, of GST pull downs of either PDZD11-HA (P11-HA) or CFP-HA (negative control) (preys, green), using as baits (red) either GST or GST fused to the indicated sequences. Bait degradation products are nonspecifically labeled in the CFP-HA pull down (red dashed rectangles). (D) IB analysis, using anti-GFP antibodies, of GST pull downs of GFP-tagged PLEKHA5 (full length), PLEKHA6 (full length), and PLEKHA7 (N-ter, 1-562) preys using as baits either GST or GST-PDZD11. (E) IB analysis, using anti-GFP antibodies, of GST pull downs of GFP-tagged PLEKHA5 (full length), PLEKHA6 (full length), and PLEKHA7 (N-ter, 1-562) preys (green) using as baits (red) either GST, or GST-PDZD11 full length, or the N-terminal 30 residues (P11-Nterm), or a mutant PDZD11 lacking the first 24 residues (P11-Δ24). Ponceau S-stained blots show baits (dashed black rectangles).
We generated antibodies against the C-terminal regions of PLEKHA5 and PLEKHA6 (Supplemental Figure S1A, orange boxes) and validated their specificity by immunoblotting (IB) and immunofluorescence (IF) microscopy analysis of cells either overexpressing (Supplemental Figure S2, A–C) or lacking (Supplemental Figure S2, D–F) the respective proteins. PDZD11 was detected in immunoprecipitates of endogenous PLEKHA5 and PLEKHA6 present in Caco-2 cells, which are of human origin, as the proteins used in the Y2H assay, indicating the formation of a complex in cells (Figure 1B). Moreover, in agreement with the results of the Y2H screen, glutathione-S-transferase (GST)-fusion proteins comprising the tandem N-terminal WW domains, but not the C-terminal domains of PLEKHA5 and PLEKHA6, interacted with full-length PDZD11 (Figure 1C). Finally, full-length PLEKHA5 and PLEKHA6 interacted with GST baits comprising either full-length PDZD11 (Figure 1D) or the first 30 residues of PDZD11, but not with PDZD11 lacking the first 24 residues (Figure 1E), demonstrating that the WW domains of PLEKHA5 and PLEKHA6 interact with the N-terminal proline-rich region of PDZD11, similarly to PLEKHA7 (Rouaud et al., 2020).
PLEKHA5, PLEKHA6, and PLEKHA7 show distinct localizations in cells and tissues and define cytoplasmic and microtubule-associated, lateral and junctional pools of PDZD11 in cultured cells
The cellular functions and localizations of PLEKHA5 and PLEKHA6 are not known. To determine whether the pattern of distribution of PLEKHA5 and PLEKHA6 is consistent with a role in ATP7A trafficking, we first examined their expression and localization in cells and tissues.
IB analysis showed distinct patterns of expression for each WW-PLEKHA in epithelial and non-epithelial cell lines (Supplemental Figure S3A and Vasileva et al., 2017). For IF microscopy analysis of endogenous proteins, we used epithelial cells from the collecting duct (mCCD) and the proximal tubule (Madin–Darby canine kidney [MDCK]) of the kidney, grown to full polarization either on Transwell filters or as cysts in Matrigel, and myeloblastic-derived Hap1 cells (Shah et al., 2018). mCCD cells express PLEKHA7 and PLEKHA6, while PLEKHA5 is not detectable by IB (Supplemental Figure S3A). In polarized mCCD cells, PLEKHA6 labeling was detected both at apical junctions, colocalizing with PLEKHA7, and along E-cadherin–labeled lateral contacts (Figure 2A). Exogenously expressed GFP-tagged PLEKHA6 and PLEKHA7 are also detected at cell–cell contacts in mCCD cells and in bEnd.3 endothelial cells, while exogenous PLEKHA5 shows in addition a cytoplasmic fibrillar staining (Supplemental Figure S3, B and C). Coexpression of PDZD11-HA with GFP-tagged PLEKHA6 and PLEKHA7 in polarized mCCD cells enhanced the accumulation of PDZD11 and its WW-PLEKHA interactor either along lateral contacts (PLEKHA6, Figure 2B) or at apical junctions (PLEKHA7, Figure 2C). Coexpression of PDZD11 with PLEKHA5 resulted in enhanced localization of both proteins in the cytoplasm and along lateral contacts (Figure 2D), whereas expression of PDZD11 alone resulted in its detection at apical junctions and lateral contacts and in the cytoplasm of mCCD cells (Figure 2E).
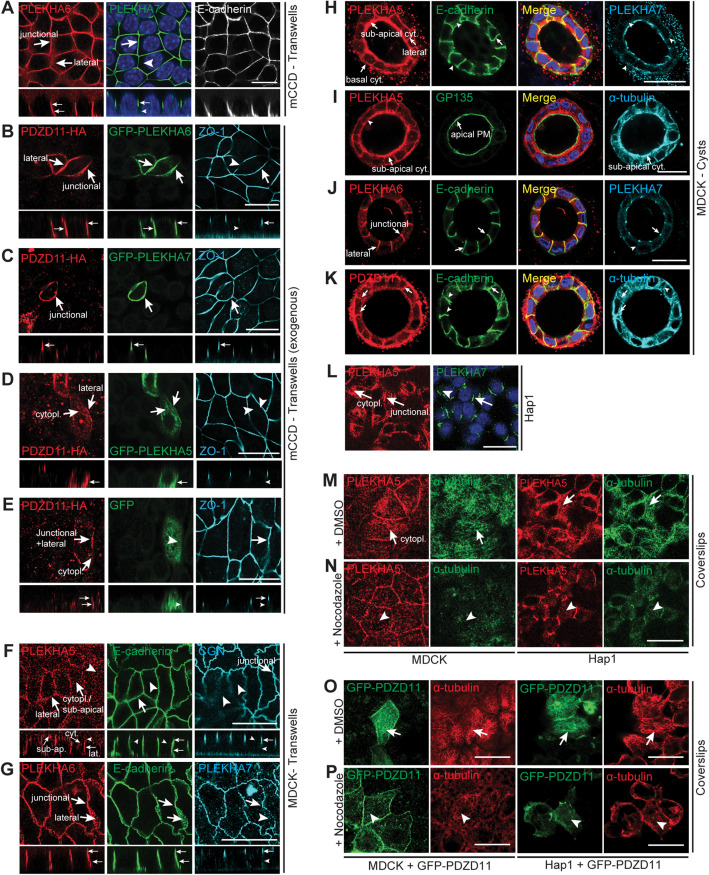
FIGURE 2:
PLEKHA5 and PLEKHA6 are localized along the basal, apical, and lateral plasma membranes in cultured cells and recruit PDZD11. (A, F–L) Immunofluorescence (IF) microscopy of endogenous PLEKHA5, PLEKHA6, and PLEKHA7 in cultured cells, using either zonular markers (ZO-1, CGN) or the AJ/lateral marker E-cadherin, or the apical marker GP135, or the microtubule protein α-tubulin as references. Different cell types and supports were used, as indicated: (A) mCCD (Transwells); (F, G) MDCK (Transwells); (H–K) MDCK (cysts in Matrigel); (L) Hap1 (coverslips). (B–E) IF microscopy of exogenous PDZD11-HA and GFP-tagged PLEKHA6 (B), PLEKHA7 (C), PLEKHA5 (D), and GFP alone (E, negative control) in mCCD cells (Transwells). (M–P) IF microscopy of endogenous PLEKHA5 (M, N) or exogenous GFP-tagged PDZD11 (O, P) in MDCK or Hap1 cells grown on coverslips, treated with either DMSO (M, O) or nocodazole (N, P), using antibodies against α-tubulin to label microtubules. For cells on Transwells (A–G), Z sections taken at the horizontal middle positions are shown below XY images. Arrows indicate labeling, arrowheads indicate low/undetectable labeling. Junctional, basal, apical plasma membrane (PM), cytoplasmic (cyt/cytopl.), subapical cytoplasmic (cyt.), and lateral (lat.) localizations/pools are indicated by arrows. Bars = 20 µm.
MDCK cells express all three WW-PLEKHAs as detected by IB (Supplemental Figure S3A and Pulimeno et al. [2010] for what concerns PLEKHA7). In MDCK cells grown on Transwells, PLEKHA5 colocalized with E-cadherin at lateral contacts, in the submembrane cytoplasm near the apical and basal domains, and in the cytoplasm, but not at junctions (Figure 2F, arrows and arrowheads), whereas PLEKHA6 was localized both at apical junctions, colocalizing with PLEKHA7, and along lateral contacts, colocalized with E-cadherin, but not in the cytoplasm (Figure 2G). In MDCK cysts, PLEKHA5 was detected along the lateral membrane, colocalizing with E-cadherin, and in the cytoplasm near basal and apical plasma membranes (Figure 2H, arrows). PLEKHA5 apical labeling was spatially distinct from that of the apical membrane marker GP135 but overlapped with tubulin labeling (Figure 2I, arrows), indicating an association with apical submembrane structures associated with microtubules, rather than a juxtamembrane localization. PLEKHA6 was localized at junctions and along the lateral membrane (Figure 2J), and PDZD11 was detected at junctions and lateral cell–cell contacts and in the cytoplasm near basal and apical membranes, overlapping tubulin staining (Figure 2K, arrows) similarly to PLEKHA5. Hap1 cells express PLEKHA5 and PLEKHA7, as detected by IB (Supplemental Figure S3A). In Hap1 cells, PLEKHA5 was colocalized with PLEKHA7 at junctions (Shah et al., 2018) and was also distributed extensively along the plasma membrane and in a cytoplasmic fibrillar pattern (Figure 2L, arrows and arrowheads). Because the localizations of PLEKHA5 and PDZD11 were similar to that of microtubules, we asked whether the integrity of the microtubule network controls PLEKHA5 localization. The cytoplasmic labeling of PLEKHA5 in both MDCK and Hap1 cells was dramatically reduced by treatment with the microtubule-depolymerizing drug nocodazole (Figure 2, M and N), indicating that cytoplasmic PLEKHA5 is associated with microtubules. Similarly, cytoplasmic labeling for exogenous PDZD11 was detected in MDCK and in Hap1 cells (Figure 2O, arrows) and was dramatically decreased upon treatment with nocodazole (Figure 2P, arrowheads).
Next, we examined the expression and localization of WW-PLEKHAs in tissues. IB analysis showed expression of WW-PLEKHAs in different tissues and faster- and slower-migrating forms, suggesting the expression of differentially spliced isoforms and/or posttranslational modifications (see also Pulimeno et al., 2010) (Supplemental Figure S3D). IF microscopy analysis of the kidney cortex showed strong PLEKHA5 labeling of glomeruli and weak diffuse labeling of apical and basal regions of tubular cells (Supplemental Figure S3E, top panels, arrows and arrowheads), whereas PLEKHA6 labeling was localized along the basal surface of tubular epithelial cells, and PLEKHA7 localized at epithelial apical junctions (Supplemental Figure S3E, bottom panels, arrows and arrowheads) (Pulimeno et al., 2010). In intestinal epithelial cells of the duodenum, PLEKHA5 was detected mostly in a subapical diffuse localization (Supplemental Figure S3F, top panel), whereas PLEKHA6 was detected both at junctions, colocalizing with PLEKHA7, and along basolateral surfaces (Supplemental Figure S3F, bottom panel, arrows). The localizations of PLEKHA5 and PLEKHA6 in intestinal epithelial cells were distinct from the localization of ATP7A, which is localized in the TGN (Supplemental Figure S3G) (Monty et al., 2005; Nyasae et al., 2007). We also tested whether WW-PLEKHAs are expressed in ATP7A-expressing cells in the brain. Strong expression of PLEKHA5 and PLEKHA7 was detected in blood vessels, marked by the endothelial marker PECAM-1 (Supplemental Figure S3H). ATP7A is highly expressed in pia matter adjacent to glial end-feet labeled with GFAP and in the outer layer of the blood vessel (Supplemental Figure S3H, green). On the contrary, PLEKHA6 is not detected in brain blood vessel (Supplemental Figure S3H, bottom panels). Expression of PLEKHA7 was broader than that of PLEKHA5, and in some cases, ATP7A colocalized with PECAM-1 and PLEKHA7 (Supplemental Figure S3H). The locus coeruleus (LC) contains the highest amount of copper among brain neurons, and ATP7A plays an important role in metalate dopamine-β-hydroxylase activity to promote norepinephrine biosynthesis (Schmidt et al., 2018; Xiao et al., 2018). We tested whether PDZD11 and WW-PLEKHAs are coexpressed in these neurons. WW-PLEKHAs and PDZD11 were detected in LC neurons; however, ATP7A-labeled puncta did not overlap with WW-PLEKHAs and showed partial colocalization with PDZD11 in LC (Supplemental Figure S3I). Overall, PLEKHA7 distribution was broader than that of both PLEKHA5 and PLEKHA6, because labeling was detected in some neurons, brain blood vessels, and choroid plexus and shows a better overlap with ATP7A expression. On the other hand, although PLEKHA5 was immunolocalized in primary hippocampal neurons (Bayes et al., 2011; Pandya et al., 2017), our results on brain sections suggest a more abundant distribution in blood vessels. Finally, in primary cultures of cortical neurons, PLEKHA5 and PLEKHA6 labeling was detected in the cytoplasm of the neuronal cell body and was colocalized with β-tubulin III along projections, whereas weak PLEKHA7 labeling was detected in the cytoplasm and nucleus (Supplemental Figure S3J, arrows). In summary, WW-PLEKHAs show different patterns of subcellular localization and tissue expression in cells and tissues that express ATP7A and do not colocalize with ATP7A under basal copper conditions.
PDZD11 and WW-PLEKHAs are required for ATP7A localization at the cell periphery in response to elevated copper
To study the role of WW-PLEKHAs in the localization and trafficking of ATP7A, we used available and new knockout (KO) cell lines for either one or two WW-PLEKHAs, or for PDZD11, in the background of mCCD (Supplemental Figure S4, A–C), MDCK (Supplemental Figure S4, D–G), and Hap1 (Supplemental Figure S4, H–J) cells. The KO of one WW-PLEKHA did not affect either the localization or levels of expression of the remaining WW-PLEKHA(s) (Supplemental Figure S5, A, B, D, and F). Moreover, KO of either PLEKHA5 or PLEKHA6 did not affect the localization of different components of junctions and lateral contacts, including nectin-3, ADAM10, and Tspan33, which interact with the PLEKHA7-PDZD11 complex (Supplemental Figure S5, A, C, and E) (Guerrera et al., 2016; Shah et al., 2018), or Tspan15, which is localized laterally in epithelial cells (Supplemental Figure S5, C and E) (Shah et al., 2018).
WT and KO clonal lines were cultured either as cysts in Matrigel or on Transwell filters, which allows optimal apicobasal polarization. In WT mCCD cells at basal copper levels, ATP7A labeling was concentrated in a perinuclear location facing the apical membrane (apical perinuclear region), previously identified as the TGN (Greenough et al., 2004; Monty et al., 2005; Nyasae et al., 2007) (Figure 3, A and C, and Supplemental Figure S6E). The same localization was observed at basal copper levels in mCCD cells KO for either PDZD11, or PLEKHA6, or PLEKHA7, or for both PLEKHA6 and PLEKHA7 (Figure 3, A and C, and Supplemental Figure S6, A, C, and F–I). The TGN localization of ATP7A was confirmed by colocalization of ATP7A with the TGN marker golgin-97 (Supplemental Figure S6, O–S, quantification in Y), and it was similar in WT and KO lines. Similarly, in both cells grown on Transwells and cysts under basal copper conditions, a perinuclear apical localization of ATP7A was observed in WT MDCK cells and in cells KO for either PLEKHA5 or PLEKHA6 (Figure 4, A and C, and Supplemental Figure S7, F–H). Moreover, colocalization between ATP7A and golgin-97 was observed in basal copper conditions in MDCK cysts (Supplemental Figure S7A) and cells grown on Transwells (Supplemental Figure S7, L–N, quantification in R) in an indistinguishable manner in WT, PLEKHA5-KO, and PLEKHA-6-KO cells, confirming the established localization of ATP7A in the TGN (Camakaris et al., 1995; Petris et al., 1996; Petris and Mercer, 1999; Yamaguchi et al., 1996; Dierick et al., 1997; La Fontaine et al., 1998). This indicates that neither PDZD11 nor WW-PLEKHAs are required for the localization of ATP7A in the TGN at basal copper levels in epithelial cells.
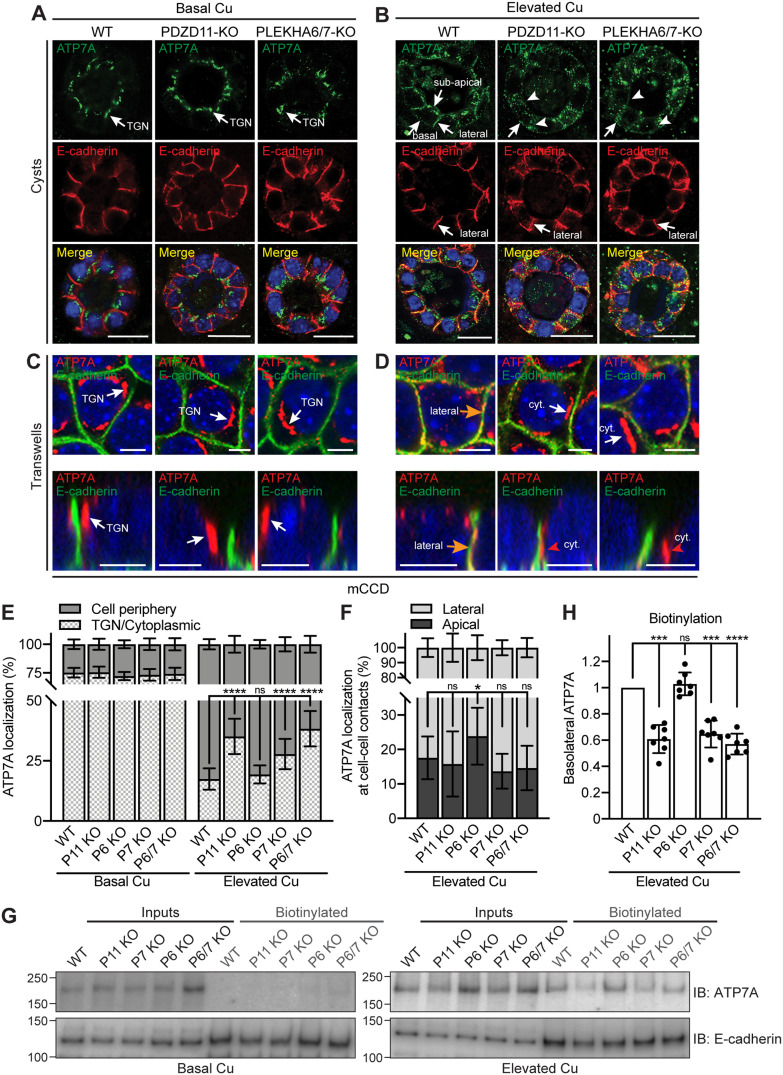
FIGURE 3:
PDZD11, PLEKHA6, and PLEKHA7 are required for the correct targeting of ATP7A to the cell periphery of mCCD cells in elevated copper. (A, B) IF microscopy of the localization of ATP7A (green) and E-cadherin (basolateral labeling marker, red) in mCCD cysts. TGN = trans-Golgi network. Lateral, subapical, and basal ATP7A labeling in elevated copper are indicated by arrows in B. Arrowheads indicate low/undetectable labeling. Bars = 20 µm. (C–F) IF microscopy (C, D) and quantifications (E, F) of ATP7A localization in mCCD cells grown on Transwells (see Supplemental Figure S6 for images at lower magnification). Orange arrows and red arrowheads in D indicate ATP7A labeling colocalized and noncolocalized with E-cadherin, respectively. Bars = 5 µm. Values of ATP7A distribution between cell periphery and TGN/cytoplasm (E) or between lateral and apical cell–cell contacts (F) are shown as mean ± SD. n = 22–30 cells for Basal Cu and n = 92–107 cells for Elevated Cu (E), n = 18–23 cell–cell contacts (F) (one-way ANOVA with post hoc Dunnett’s test, *p < 0.05, ****p < 0.0001, ns, not significant). (G) IB analysis of biotinylated ATP7A in the indicated WT and KO lines under basal and high copper conditions. E-cadherin was used as positive control for basolateral biotinylation. (H) Quantification of basolateral ATP7A in Elevated Cu indicates for each genotype the biotinylated ATP7A signal normalized to the input, relative to WT cells. Dots show replicates (n = 7), and bars represent mean ± SD. One-way ANOVA with post hoc Dunnett’s test (***p < 0.001, ****p < 0.0001, ns, not significant).
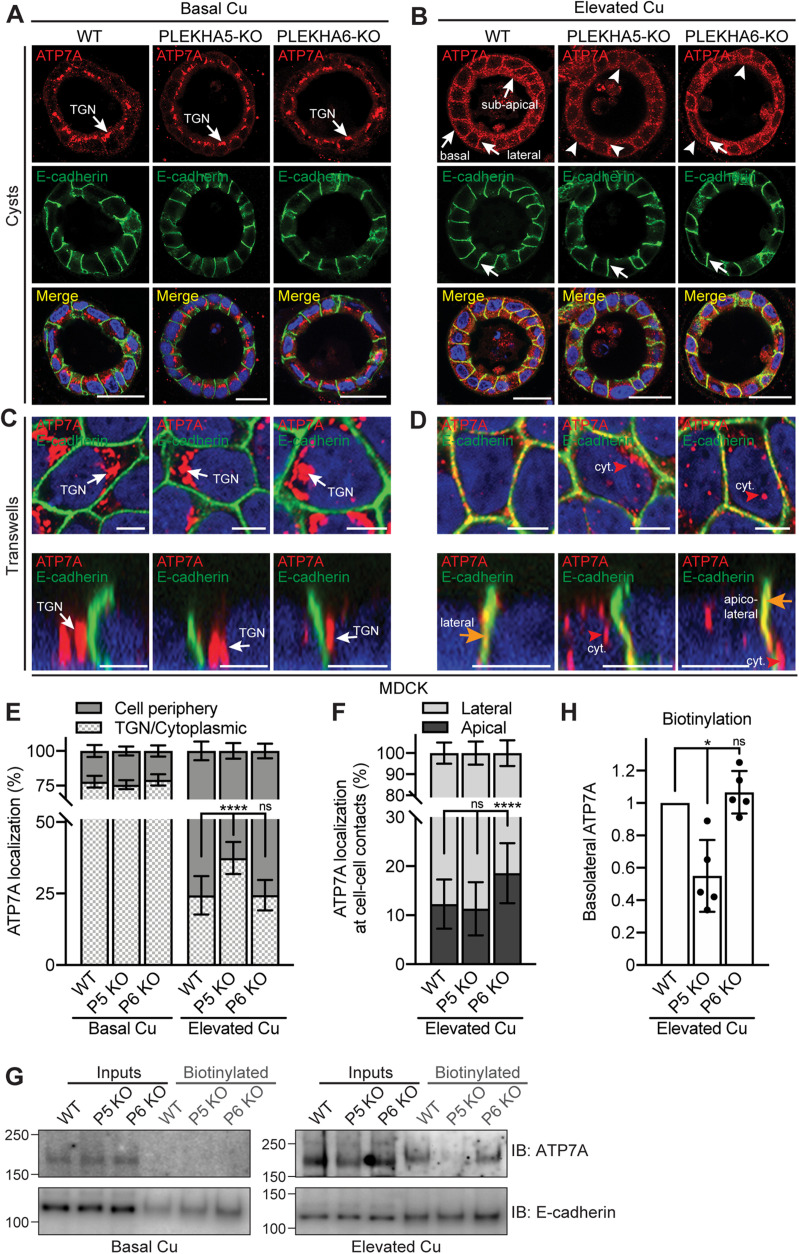
FIGURE 4:
PLEKHA5 and PLEKHA6 are required for the correct targeting of ATP7A to the cell periphery of MDCK cells in elevated copper. (A, B) IF microscopy of the localization of ATP7A in MDCK cysts. TGN = trans-Golgi network. Lateral, subapical, and basal ATP7A labeling in elevated copper is indicated by arrows in B. Arrowheads indicate low/undetectable labeling. Bars = 20 µm. (C–F) IF microscopy (C, D) and quantifications (E, F) of ATP7A localization in MDCK cells grown on Transwells (see Supplemental Figure S7 for images at lower magnification). Orange arrows and red arrowhead in D indicate ATP7A labeling colocalized and noncolocalized with E-cadherin, respectively. Bars = 5 µm. Values of ATP7A distribution between cell periphery and TGN/cytoplasm (E) or between lateral and apical cell–cell contacts (F) are shown as mean ± SD. n = 25–31 (E, Basal Cu) and 90–105 cells (E, Elevated Cu), n = 29–47 cell–cell contacts (F). One-way ANOVA with post hoc Dunnett’s test (****p < 0.0001, ns, not significant). (G) IB analysis of biotinylated ATP7A in the indicated WT and KO lines under basal and high copper conditions. E-cadherin was used as positive control for basolateral labeling. (H) Quantification of basolateral ATP7A in Elevated Cu indicates for each genotype the biotinylated ATP7A signal normalized to the input, relative to WT cells. Dots show replicates (n = 5), and bars represent mean ± SD. One-way ANOVA with post hoc Dunnett’s test (*p < 0.05, ns, not significant).
Upon exposure to elevated copper levels, WT mCCD cells in cysts showed a massive redistribution of ATP7A labeling from the TGN to the cell periphery. Strong ATP7A labeling was detected linearly along lateral contacts and basal plasma membranes and in a subapical position (Figure 3B, WT, arrows). Instead, in mCCD cysts KO either for PDZD11 or for both PLEKHA6 and PLEKHA7, accumulation of ATP7A near the plasma membranes was disrupted (Figure 3B, PDZD11-KO and PLEKHA6/7-KO, arrowheads). In mCCD cysts KO for PLEKHA6 alone, the lateral accumulation of ATP7A was similar to that of WT cysts, whereas in cysts KO for PLEKHA7 alone the submembrane labeling was disrupted (Supplemental Figure S6B). In WT mCCD cells grown on Transwells, TGN labeling for ATP7A strongly decreased in elevated copper, whereas labeling proximal to lateral E-cadherin was strongly increased (Figure 3D, orange arrows; Supplemental Figure S6J). Instead, cells KO for either PDZD11, or both PLEKHA6 and PLEKHA7, or PLEKHA7 alone showed decreased ATP7A labeling at the cell periphery along E-cadherin–labeled lateral contacts and increased cytoplasmic perinuclear staining (Figure 3D, cyt. and red arrowheads; Supplemental Figure S6, D, K, M, and N, quantification in Figure 3E). It should be noted that some ATP7A labeling was still detected in the TGN of WT and KO cells, colocalized with golgin-97 in elevated copper conditions, albeit less than in basal copper (Supplemental Figure S6, T–X, quantification in Z). In mCCD cells KO for PLEKHA6, the redistribution of ATP7A to the cell periphery was similar to that in WT cells (Supplemental Figure S6, D and L, quantification in Figure 3E), but labeling for ATP7A was shifted toward apicolateral junctional contacts, rather than lateral cell–cell contacts (quantification in Figure 3F).
In WT MDCK cysts exposed to elevated copper, strong ATP7A labeling was detected along lateral contacts, near the basal plasma membrane, and in a subapical localization, similarly to mCCD cells (Figure 4B, WT, arrows). Double IF microscopy analysis with the apical membrane marker GP135 confirmed that ATP7A is in a subapical, and not apical, localization (Supplemental Figure S7, C and D). The KO of PLEKHA5 resulted in a strong decrease in subapical, basal, and lateral ATP7A labeling in elevated copper, compared with that in WT cysts (Figure 4B, arrowheads, PLEKHA5-KO). The KO of PLEKHA6 resulted instead in disorganized and reduced subapical and basal labeling, but not as strong as that observed in PLEKHA5-KO cysts (Figure 4B, PLEKHA6-KO, arrow and arrowheads). Some residual signal for ATP7A was observed in the TGN of MDCK cells grown in cysts, with no difference between WT and KO lines (Supplemental Figure S7B). Immunofluorescence microscopy analysis of ATP7A and the early sorting endosomal marker EEA1 in MDCK cysts in elevated copper conditions showed that ATP7A labeling is distinct from that of EEA1 (Supplemental Figure S7E), suggesting that ATP7A-associated vesicles are not early sorting endosomes.
In MDCK cells grown on Transwells and exposed to elevated copper levels, KO of PLEKHA5 resulted in a significant increase in ATP7A labeling in the cytoplasmic space between the perinuclear TGN and the cell periphery, and in PLEKHA6-KO cells the redistribution of ATP7A to the cell periphery was similar to that in WT cells (Figure 4D and Supplemental Figure S7, I–K, quantifications in Figure 4E). ATP7A labeling was mostly detected in the basolateral and subapical cell periphery, with no significant ATP7A staining colocalized with golgin-97 in the TGN in both WT and KO cells (Supplemental Figure S7, O–Q, quantification in S), confirming that WW-PLEKHAs do not modulate the exit of ATP7A from the TGN, but rather its association with the cell periphery. As in cysts, no colocalization was observed between ATP7A and the early sorting endosomal marker EEA1 in MDCK cells grown on Transwells and exposed to elevated copper conditions, either WT or PLEKHA5-KO or PLEKHA6-KO (Supplemental Figure S7, T–V). Interestingly, in cells KO for PLEKHA6, lateral labeling for ATP7A was shifted apically (Figure 4D, quantification in F).
Next, we asked whether either PDZD11 or WW-PLEKHAs are required for the retrograde trafficking of ATP7A to the TGN when the elevated copper levels are depleted by treatment with the copper chelator bathocuproinedisulfonic acid (BCS). IF microscopy analysis revealed that both mCCD (Supplemental Figure S6, AA–AE) and MDCK (Supplemental Figure S7, W–Y) WT and KO lines showed the same redistribution of ATP7A back to the TGN region after copper washout, with ATP7A colocalizing with golgin-97 (Supplemental Figure S6, AF–AK, for mCCD and Supplemental Figure S7, Z–AC, for MDCK).
Finally, because previous work showed that copper induces increased basolateral plasma membrane levels of ATP7A, as measured by biotinylation (Greenough et al., 2004), we examined the role of WW-PLEKHAs and PDZD11 in modulating the surface levels of ATP7A along the basolateral membrane. Basolateral surface proteins were biotinylated by the addition of sulfo-NHS-SS-biotin to the basal chamber, isolated by affinity chromatography on streptavidin-coated beads, and lysates were analyzed by SDS–PAGE and IB using anti-ATP7A antibodies and anti–E-cadherin antibodies as a positive control (Figure 3G for mCCD and Figure 4G for MDCK). Apical biotinylation was not carried out because previous biotinylation studies (Greenough et al., 2004; Nyasae et al., 2007) demonstrated that the amount of ATP7A at the apical surface is negligible and not physiologically relevant. Our data also show that in our cellular models, ATP7A is not localized at the apical membrane, but in a subapical localization even in elevated copper (Supplemental Figure S7D). No basolateral ATP7A was detected under basal copper conditions, whereas elevated copper resulted in detectable ATP7A (Figure 3G for mCCD and Figure 4G for MDCK), in agreement with Greenough et al. (2004). Confirming our IF microscopy analysis, KO of either PDZD11, or PLEKHA7, or both PLEKHA6 and PLEKHA7, but not PLEKHA6 alone, resulted in decreased levels of ATP7A at the basolateral surface of mCCD cells, when compared with WT (Figure 3G, quantifications in H). In MDCK cells, we observed a decrease in the basolateral levels of ATP7A in PLEKHA5-KO, but not PLEKHA6-KO cells, when compared with WT (Figure 4G, quantification in H).
In summary, these findings indicate that PDZD11 and WW-PLEKHAs are not required either for the TGN localization of ATP7A under basal copper conditions, or for the copper-induced exit from the TGN, or for the retrograde traffic of ATP7A to the TGN after copper washout. However, they are required to promote the efficient localization of ATP7A at the cell periphery.
PDZD11 and WW-PLEKHAs regulate intracellular copper homeostasis in response to elevated copper level
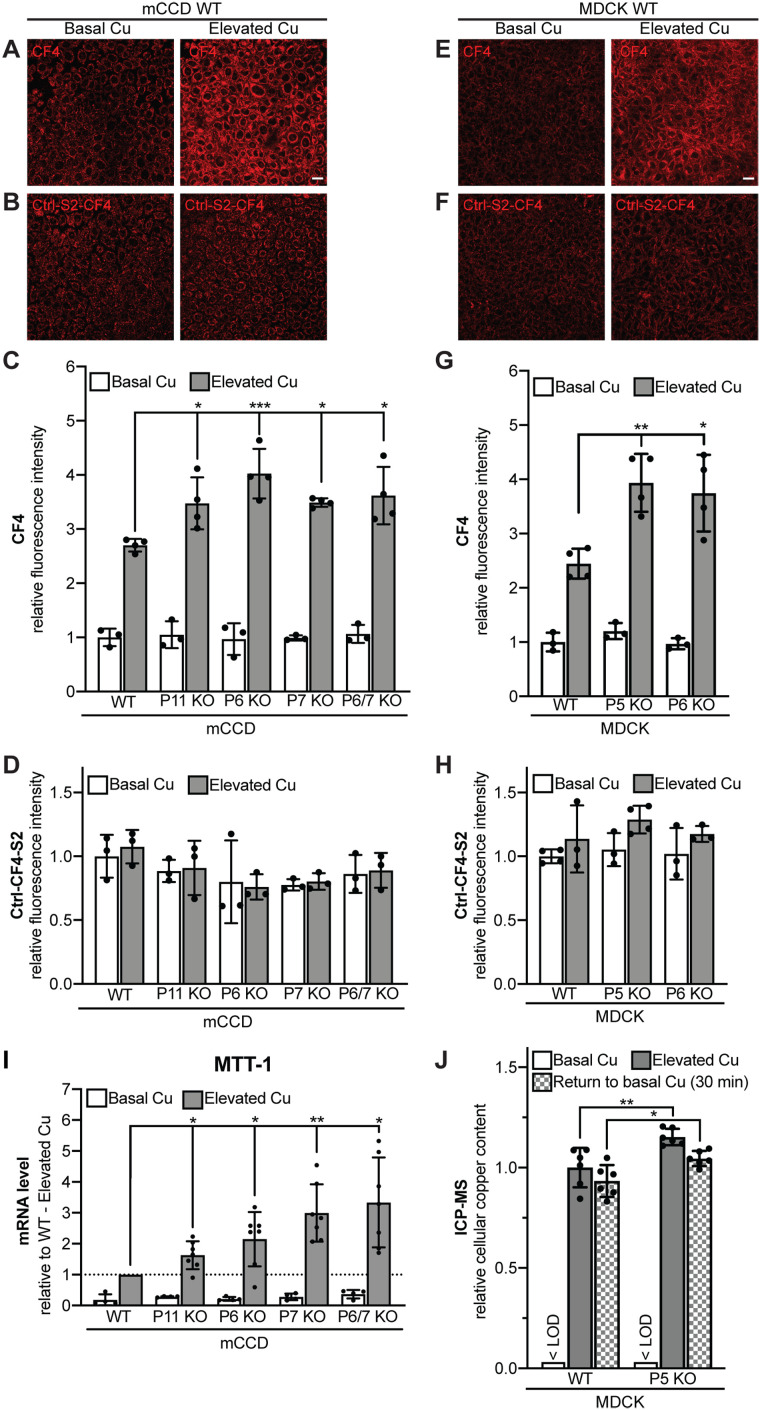
We asked whether the altered localization of ATP7A in cells KO for either PDZD11 or WW-PLEKHAs correlates with changes in copper homeostasis. For this purpose, we first used the Copper Fluor-4 (CF4) probe (Figure 5, A and E), which, in combination with the control Copper Fluor-4-Sulfur-2 (Ctrl-CF4-S2) sensor that is insensitive to copper changes (Figure 5, B and F), provides a measure of intracellular labile copper (Xiao et al., 2018).
FIGURE 5:
PDZD11 and WW-PLEKHAs are required for the control of labile copper in mCCD and MDCK cells exposed to elevated copper level. Fluorescence images (A, B, E, F) (scale bar = 20 µm), and quantification of fluorescence (C, D, G, H) of either mCCD (A–D) or MDCK (E–H) cells loaded either with CF4 (A, C, E, G) or Ctrl-CF4-S2 (B, D, F, H) under conditions of basal copper (Basal Cu) and after treatment with high concentration of copper (Elevated Cu). In C, D, G, and H, dots show replicates (n), relative to the mean of WT cells in basal copper, and bars represent mean ± SD. One-way ANOVA with post hoc Dunnett’s test (*p < 0.05, **p < 0.01, ***p < 0.001). (I) Quantification by qRT-PCR of the mRNA levels of metallothionein-I (MTT-I) (using GAPDH as internal standard, see Materials and Methods) in WT and KO mCCD cells under conditions of basal copper (Basal Cu) and after treatment with high concentration of copper (Elevated Cu), relative to elevated copper-treated WT cells. Dots show replicates (n = 4 for Basal Cu, n = 7 for Elevated Cu), and bars represent mean ± SD. One-way ANOVA with post hoc Dunnett’s test (*p < 0.05, **p < 0.01). (J) Measurement of cellular copper content by ICP-MS in WT and PLEKHA5 KO MDCK cells under conditions of basal copper (Basal Cu), after treatment with high concentration of copper (Elevated Cu) and after treatment with elevated copper followed by a 30-min treatment in Basal Cu medium (Return to basal Cu) (see Materials and Methods). < LOD = below limit of detection. Dots show replicates (n = 6), relative to the mean of WT cells in elevated copper, and bars represent mean ± SD. One-way ANOVA with post hoc Sidak test (*p < 0.05, **p < 0.01). Abbreviations for genotypes: P11 = PDZD11; P5 = PLEKHA5, P6 = PLEKHA6; P7 = PLEKHA7.
In agreement with the normal TGN localization of ATP7A, in basal copper conditions KO cells showed intracellular labile copper levels similar to those of WT cells, in both mCCD and MDCK lines (Figure 5, C and G). In contrast, exposure to elevated copper resulted in a 2.7-fold increase in intracellular labile copper in WT mCCD cells, but greater increases, between 3.4-fold and 4.1-fold, in cells KO for either PDZD11, PLEKHA6, PLEKHA7, or both PLEKHA6 and PLEKHA7 (Figure 5C). Similarly, in WT MDCK cells, elevated copper resulted in a 2.4-fold increase in intracellular labile copper, whereas in cells KO for either PLEKHA5 or PLEKHA6, the increase was 3.9-fold and 3.7-fold, respectively (Figure 5G). The absence of significant differences in Ctrl-CF4-S2 fluorescent signals between WT and KO cells, in either basal or elevated copper environments, indicated comparable loading of the probes in all conditions (Figure 5, D and H).
To confirm that PDZD11 and WW-PLEKHAs control intracellular copper levels, we measured the mRNA expression of metallothionein-1 (MTT-1), which is regulated by intracellular copper (Mercer et al., 1981), in WT and KO cells. In elevated copper conditions, the increase in MTT-1 expression was significantly higher in cells KO for either PDZD11, PLEKHA6, PLEKHA7, or both PLEKHA6 and PLEKHA7 (Figure 5I). Furthermore, quantification of total cellular copper content by ICP-MS confirmed a higher increase in copper levels in PLEKHA5-KO MDCK cells with respect to WT in elevated copper conditions and after return to basal copper conditions (Figure 5J).
Because elevated intracellular copper levels are toxic (Schilsky, 1996), we predicted that cell death would increase upon elevated copper in cells KO for either PDZD11 or WW-PLEKHAs. To test this hypothesis, we carried out a cell viability assay using crystal violet for both MDCK (Supplemental Figure S8A) and mCCD (Supplemental Figure S8C) WT and KO clones. A significant decrease in cell viability was observed in PLEKHA5-KO and PLEKHA6-KO MDCK clones (Supplemental Figure S8B) and mCCD PDZD11-KO, PLEKHA6-KO, PLEKHA7-KO, and double PLEKHA6/7-KO clones (Supplemental Figure S8D), when compared with WT cells.
Together, these results support the notion that WW-PLEKHAs and PDZD11 regulate copper homeostasis, resulting in higher intracellular copper accumulation in KO cells exposed to elevated copper levels, through impaired trafficking of ATP7A to the cell periphery.
WW-PLEKHAs promote the binding of the C-terminal region of ATP7A to PDZD11
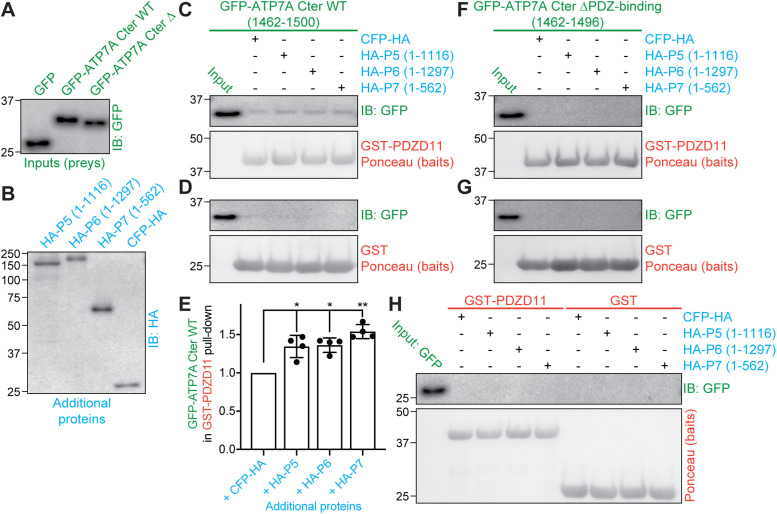
Because PDZD11 binds both to the C-terminus of ATP7A and to the WW domains of WW-PLEKHAs, we investigated whether WW-PLEKHAs modulate ATP7A-PDZD11 interaction using a trimolecular GST pull-down assay. We used as preys GFP-tagged constructs encoding either the C-terminus of ATP7A (residues 1462–1500) or the same construct lacking the PDZ-binding motif (residues 1462–1496), or GFP alone (negative control) (Figure 6A). As additional (third) proteins, we used HA-tagged constructs of PLEKHA5, PLEKHA6, and PLEKHA7 (Figure 6B). Either GST-PDZD11 (Figure 6, C and F) or GST alone (negative control; Figure 6, D and G) was used as bait. IB analysis showed that the C-terminus of ATP7A but not GFP interacted with PDZD11, and this interaction increased in the presence of WW-PLEKHAs (Figure 6, C–E). In contrast, no interaction between PDZD11 and the C-terminus of ATP7A was observed when the PDZ-binding motif of ATP7A was deleted (Figure 6, F and G). Either GST-PDZD11 or GST alone did not interact with GFP alone used as a prey (negative control; Figure 6H). These results indicate that WW-PLEKHAs promote the interaction of the PDZ-binding motif of the C-terminus of ATP7A with PDZD11.
FIGURE 6:
WW-PLEKHAs enhance the interaction of PDZD11 with the C-terminus of ATP7A. (A, B) Normalized preys (A, green) and additional proteins (B, blue) for GST pull downs. Preys normalized by IB with anti-GFP (A) were either GFP, GFP-tagged C-terminus of ATP7A WT (1465–1500), or the same, but lacking the C-terminal four-residue PDZ-binding motif (GFP-ATP7A Cter ΔPDZ-binding). Additional proteins normalized with anti-HA (B) antibodies were either full-length PLEKHA5 (P5), full-length PLEKHA6 (P6), N-terminal fragment of PLEKHA7 (P7), or CFP-HA (negative control). (C–H) GST pull downs. IB analysis using anti-GFP antibodies (C, D, F–H) and signal quantification (E) of GST pull downs using either GST or GST-PDZD11 as baits (Ponceau S staining below IB shows baits, in red), and the indicated preys (green), carried out either in the presence or the absence of the indicated HA-tagged third additional protein (blue). (E) quantifications, with each dot representing a replicate (n = 4) of densitometric analysis of the GFP immunoblot in C (pull down of GFP-ATP7A Cter WT using GST-PDZD11 as a bait under the different conditions), each normalized to the respective signal of GST-PDZD11 bait (see Materials and Methods). Bars represent mean ± SD. Repeated measures (RM) one-way ANOVA followed by Dunnett’s multiple comparison test with +CFP-HA as reference (*p < 0.05, **p < 0.01).
To verify that the interaction detected in vitro occurs within cells, we used immunofluorescence colocalization, because coimmunoprecipitation of endogenous full-length ATP7A was not possible because it required solubilization conditions that abolished labile protein–protein interactions. There was no colocalization between ATP7A and PDZD11 and ATP7A and either PLEKHA5, PLEKHA6, or PLEKHA7 under basal copper conditions (Supplemental Figure S9, A–C). In contrast, in elevated copper conditions, we observed colocalization of ATP7A labeling with PDZD11 and PLEKHA5 both at the cell periphery (Supplemental Figure S9D, magnified insets a in D′ and D′′) and in the cytoplasm (Supplemental Figure S9D, magnified insets b in D′ and D′′). We also observed colocalization of ATP7A labeling with PDZD11 and PLEKHA6 (Supplemental Figure S9E, magnified insets a in E′ and E′′) and PLEKHA7 (Supplemental Figure S9F, magnified insets a in F′ and F′′) at the cell periphery. Thus ATP7A, PDZD11, and the WW-PLEKHA proteins partially overlap in the same compartments under elevated copper conditions, and a potential physical association in vivo is indicated by the in vitro interaction between the C-terminal PDZ-binding domain of ATP7A and PDZD11, which is increased in the presence of WW-PLEKHAs.
DISCUSSION
The trafficking of ATP7A and the regulation of its localization are critical for the control of intracellular copper homeostasis, but little is known about the trafficking machinery that drives and stabilizes ATP7A at the cell periphery (Holloway et al., 2007, 2013; La Fontaine and Mercer, 2007; Lutsenko et al., 2007; Veldhuis et al., 2009; Polishchuk and Lutsenko, 2013; Skjorringe et al., 2017; Hartwig et al., 2019). Here we identify PLEKHA5, PLEKHA6, and PLEKHA7, and their ligand adaptor PDZD11, as proteins involved in the copper-dependent localization of ATP7A at the cell periphery and in the maintenance of copper homeostasis.
PLEKHA5 and PLEKHA6 were first characterized as PH domain–containing proteins implicated in phosphoinositide signaling (Dowler et al., 2000), and genetic studies indicate that they participate in several developmental processes and diseases (Wythe et al., 2011; Fromer et al., 2014; Jamain et al., 2014; Spellmann et al., 2014; Jilaveanu et al., 2015; Thapa et al., 2015; Shah et al., 2016; Barbitoff et al., 2018; Cox et al., 2018; Tavano et al., 2018; Daulagala et al., 2019; Huang et al., 2020; Liu et al., 2020). However, little is known about the cellular localization and functions of PLEKHA5 and PLEKHA6 and the molecular basis for the involvement of all three WW-PLEKHAs in physiological and pathological processes.
Here we show that the tissue distributions of PLEKHA5 and PLEKHA6 are distinct from that of PLEKHA7 and that WW-PLEKHAs and PDZD11 are expressed in essentially all cell types and tissues that express ATP7A. For example, in the adult mouse brain, ATP7A is expressed in endothelial cells of blood vessels (Qian et al., 1998), ependymal cells of the choroid plexus in ventricles (Kuo et al., 1997; Choi and Zheng, 2009), a subset of astrocytes (Kodama et al., 1991), and neurons (Iwase et al., 1996), and we detected WW-PLEKHAs in most of these cell types. PLEKHA5 appears less expressed than PLEKHA6 and PLEKHA7 in neuronal and epithelial tissues and more expressed in vascular tissues. It is likely that each tissue and cell type expresses a specific combination of WW-PLEKHA proteins, providing functional redundancy, which could explain why genetic disorders with impairment of copper metabolism have not been described following loss of function of either one WW-PLEKHA or PDZD11. Indeed, our results do not exclude that ATP7A traffic may also be regulated by PDZD11- and WW-PLEKHAs–independent mechanisms, providing an additional layer of functional redundancy.
The subcellular localizations of WW-PLEKHAs are also distinct, because PLEKHA5 is associated with microtubules and PDZD11 throughout the cytoplasm (see also Zou and Cox, 2013) and is also detected near lateral membranes and in a subapical localization, whereas PLEKHA6 is detected along basolateral membranes and at apicolateral AJ, and PLEKHA7 exclusively at apicolateral AJ. The role of the WW, PH, and coiled-coil domain of WW-PLEKHAs in directing their subcellular localization was addressed in our recent study (Sluysmans et al., 2021). In the case of PLEKHA5, the PH domain is required for membrane association, whereas the PH domain of PLEKHA7 is not required for junctional localization but promotes the junctional localization of chimeric WW-PLEKHA proteins (Sluysmans et al., 2021). Thus, it cannot be excluded that PH domains of WW-PLEKHAs are involved in creating targeting patches for ATP7A-containing vesicles at the plasma membrane.
ATP7A and WW-PLEKHAs are not colocalized under basal copper conditions in either cells or tissues, and the colocalization of ATP7A with golgin-97 under basal copper conditions confirms its localization in the TGN. The KO of either PDZD11 or WW-PLEKHAs does not significantly affect the localization of ATP7A in the TGN under basal copper and the copper-induced exit of ATP7A from the TGN. This is in agreement with the observation that ATP7A localization in the TGN in basal copper conditions depends on sequences in transmembrane domain 3 (Francis et al., 1998), and the copper-induced exit from the TGN depends on one CXXC metal-binding site at the C-terminus (Lutsenko et al., 1997; Goodyer et al., 1999; Strausak et al., 1999), but not on the C-terminal sequences that interact with PDZD11 (Greenough et al., 2004; Stephenson et al., 2005). The subcellular localization of ATP7A is very sensitive to copper status, as high bioavailable copper levels target ATP7A to the cell periphery to facilitate copper efflux (Kaler, 2011). KO of either PDZD11 or WW-PLEKHAs, especially PLEKHA5 and PLEKHA7, correlates with decreased accumulation of ATP7A at the cell periphery, as detected by both IF microscopy analysis and biotinylation experiments. In PLEKHA6-KO cells, ATP7A lateral labeling and biotinylated levels were not decreased, but lateral labeling shifted apically, suggesting that ATP7A is redundantly targeted to the lateral membrane by more than one WW-PLEKHA and that PLEKHA6 retains ATP7A laterally. Because PDZD11 is distributed in cellular pools associated with different WW-PLEKHAs, the redistribution of ATP7A toward the apical junction in PLEKHA6-KO cells may be driven by an increased proportion of PDZD11 bound to junctional PLEKHA7. The significance of the subapical ATP7A labeling detected in MDCK cysts in elevated copper is unclear, because studies on mice show a redistribution of ATP7A from the TGN to basolateral but not subapical regions of the plasma membrane in intestinal and kidney epithelial cells of mice exposed to elevated copper (Monty et al., 2005; Nyasae et al., 2007; Linz et al., 2008). Our colocalization experiments with EEA1 indicate that in elevated copper conditions ATP7A is associated with vesicles that are distinct from early sorting endosomes. Thus, it is possible that ATP7A-associated vesicles are recycling endosomes, and additional studies are required to precisely identify the molecular composition of the ATP7A-containing vesicles.
Finally, PDZD11 and WW-PLEKHAs did not control retrograde trafficking of ATP7A to the TGN after copper washout, in agreement with normal recycling of the ATP7A mutant lacking the PDZ-binding motif after restoration of basal copper levels (Greenough et al., 2004). Previous studies showed that internalization of ATP7A from the plasma membrane depends in part on clathrins and clathrin adaptors AP-1 and AP-2 and in part on clathrin-independent endocytic pathways (Holloway et al., 2013; Yi and Kaler, 2015).
Interestingly, no genetic study has so far revealed a role for WW-PLEKHAs and PDZD11 in copper metabolism in vivo. PLEKHA7-KO mice are viable and were not reported to display any phenotype consistent with grossly altered copper homeostasis (Popov et al., 2015), and mice KO for either PLEKHA5 or PLEKHA6 or PDZD11 have not been described. If WW-PLEKHAs have redundant functions in the regulation of ATP7A, simultaneous KO of multiple WW-PLEKHAs may be necessary to elicit a strong phenotype. Furthermore, we cannot exclude that additional PDZ-containing proteins may also participate in the trafficking of ATP7A, potentially compensating a pathological or experimental loss of PDZD11. Thus, future studies should use mouse models, including mouse models with copper accumulation (Atp7b KO mice) (Huster et al., 2006), to test the relevance of WW-PLEKHAs and PDZD11 as targets or effectors of copper-sensing mechanisms in specific tissues.
The altered localization of ATP7A in cells KO for PDZD11 and WW-PLEKHAs correlated with increased intracellular labile and total copper levels upon exposure to elevated copper. This could be simply explained by the reduced trafficking of ATP7A-containing vesicles to the plasma membrane. However, we did not observe a strict correlation between the effect of KO on ATP7A localization and the increase in labile copper signal. This suggests that WW-PLEKHAs may also regulate the activity of ATP7A independently of their effect on ATP7A localization, for example by modulating the recycling and dynamics of ATP7A-containing vesicles (Petris et al., 1996; La Fontaine et al., 1998; Pase et al., 2004), by tethering vesicles to specific domains of the plasma membrane, or by maintaining a conformation of ATP7A that maximizes its function as a copper pump. It should be noted that the trafficking of ATP7A depends on actin and microtubule cytoskeletons (Cobbold et al., 2002, 2004), and PLEKHA7 was identified as a GTPase-activating protein (GAP) for Rac1 and Cdc42 (Lee et al., 2017) and an indirect linker to microtubules (Meng et al., 2008), and we showed that PLEKHA5 associates with microtubules (see also Zou and Cox, 2013). Thus, it is also possible that WW-PLEKHAs regulate ATP7A trafficking by affecting the dynamics of the cytoskeleton. These questions, the identification of additional components of the trafficking machinery for ATP7A, and the role of regulation by phosphorylation should be addressed by future studies.
Here we did not analyze the role of WW-PLEKHAs and PDZD11 in the regulation of the Wilson copper pump (ATP7B), because, unlike ATP7A, ATP7B lacks the PDZ-binding motif that interacts with PDZD11, and its C-terminus does not interact with PDZD11 (AIPP1) by β-galactosidase assays in yeast (Stephenson et al., 2005). Although ATP7B is not expected to be regulated by WW-PLEKHAs through PDZD11, future studies should address the localization of ATP7B in cells KO for WW-PLEKHAs, because we cannot rule out that alternatively spliced isoforms of PLEKHA proteins that lack the WW domains could be involved in the trafficking of both ATP7A and ATP7B, through PDZD11-independent mechanisms.
We noted that WW-PLEKHAs promote the interaction of PDZD11 with the PDZ-binding motif of ATP7A. These results are in line with the idea that by interacting together, WW-PLEKHAs and PDZD11 cooperatively promote the binding of the complex to their ligands (Guerrera et al., 2016; Shah et al., 2018; Rouaud et al., 2020). The WW domains of PLEKHA5 and PLEKHA6 interact with the N-terminal, proline-rich sequences of PDZD11, similarly to PLEKHA7 (Guerrera et al., 2016; Rouaud et al., 2020), and the PDZ-binding motif of ATP7A is required for binding of the ATP7A C-terminus to the PDZD11-WW-PLEKHA complex. Thus, the mode of interaction of the WW-PLEKHA-PDZD11 complex with ATP7A is similar to what was described for the Ig-like adhesion molecules nectins (Guerrera et al., 2016) and distinct from that of Tspan33, which interacts directly with the first WW domain of PLEKHA7 (Rouaud et al., 2020). Our colocalization analysis also suggests that multimolecular complexes comprising ATP7A, PDZD11, and all WW-PLEKHAs can occur at the cell periphery in elevated copper, and complexes of ATP7A, PDZD11, and PLEKHA5 can be associated with microtubules in the cytoplasm.
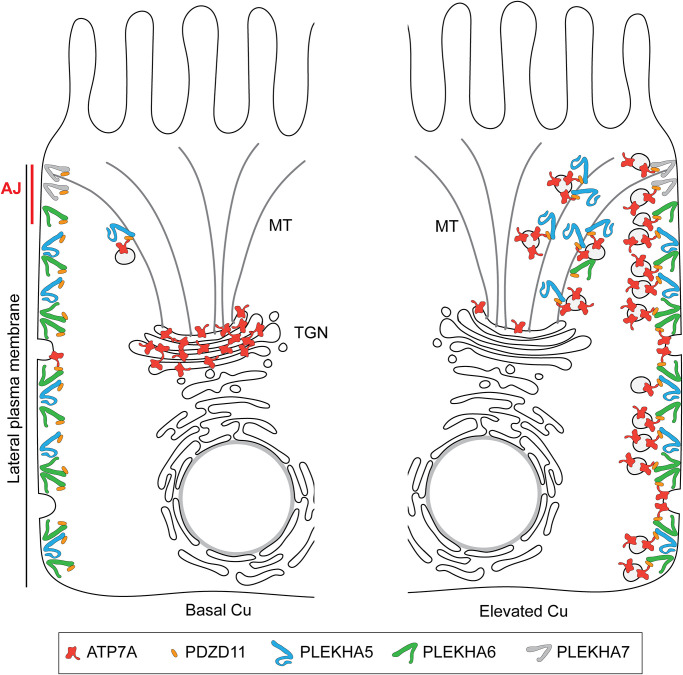
Collectively, our results indicate that PDZD11 links WW-PLEKHAs to the C-terminus of ATP7A and suggest that the WW-PLEKHA-PDZD11 complexes are involved in the trafficking, membrane delivery, tethering, and regulation of dynamic exocytosis and endocytosis of ATP7A-containing vesicles at the cell periphery in elevated copper (Figure 7). How elevated copper acts as a transition metal signal (Chang, 2015; Ackerman and Chang, 2018) to trigger the formation of the multimolecular ATP7A-PDZD11-WW-PLEKHA complexes remains to be resolved.
FIGURE 7:
Model WW-PLEKHA- and PDZD11-regulated ATP7A trafficking. Schematic model and graphical legend describing the localization of the indicated proteins and the implication of WW-PLEKHAs and PDZD11 in the anterograde trafficking of ATP7A in absorptive polarized epithelial cells (MDCK) either in basal Cu (left) or in elevated Cu (right), based on IF results shown in Figures 3 and 4 and Supplemental Figures S6 and S7. The lateral plasma membrane (PM) and the apico-lateral AJ are shown on the left. PLEKHA7 is exclusively localized at the AJ/ZA. Under basal copper (Basal Cu, left), few ATP7A-containing membrane vesicles cycle between the plasma membrane and trans-Golgi network (TGN), and most ATP7A is in the TGN. With elevated copper (Elevated Cu, right), ATP7A-containing membrane vesicles are trafficked to the cell periphery along microtubule (MT) tracks and are tethered to the apical lateral and basal PM by the WW-PLEKHA-PDZD11 complexes. Copper chaperones, transporters, adaptors, and signaling proteins involved in copper homeostasis (La Fontaine and Mercer, 2007; Lutsenko et al., 2007; Polishchuk and Lutsenko, 2013) are not shown for the sake of simplicity.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
Culture conditions for mouse cortical collecting duct cells (mCCD), MDCK cells (MDCKII Tet-off), the mouse brain microvascular endothelial (endothelioma) cell line (bEnd.3), the mouse heart endothelial cell line (H5V), the human lung carcinoma cell line (A427), ciliated aortic mouse embryonic endothelial cells (meEC), the human keratinocyte cell line (HaCaT) (Vasileva et al., 2017), human umbilical vascular endothelial cells (HUVEC) (Rouaud et al., 2019), haploid human cells (Hap1) (Popov et al., 2015), mouse mammary epithelial cells (Eph4), human intestinal carcinoma cells (Caco-2) (Spadaro et al., 2017), and human embryonic kidney epithelial cells (HEK293T) (Rouaud et al., 2020) were described previously.
Cysts of mCCD and MDCK cells were obtained using the protocol of Debnath et al. (2003). Matrigel (40 µl) (BD Biosciences; 354230) was added on glass coverslips in a 24-well plate and allowed to solidify for 30 min at 37°C. Cells were trypsinized, resuspended in SMEM (Sigma-Aldrich; M8167), pelleted by centrifugation (150 × g, 3 min), and resuspended in 2 ml of SMEM to obtain a single-cell suspension. Cells were diluted to obtain 35,000 cells/ml and mixed in a 1:1 ratio with assay medium 2×/Matrigel 4%/epidermal growth factor (EGF) (10 ng/ml), and 400 µl was plated per well. MDCK cysts were grown up to 7 d and mCCD up to 14 d, replacing medium with fresh assay medium (1×) every 4 d.
Primary cultures of cortical neurons were obtained as described in Chassefeyre et al. (2015). Cortices were dissected from E18.5 mouse embryos in Hanks’ balanced salt solution (HBSS; Invitrogen) containing HEPES 10 mM, streptomycin 10 µg/ml, penicillin 10 U/ml, then treated with 0.25% trypsin-EDTA for 10 min at 37°C and disrupted by 10 aspirations/ejections through a 1-ml micropipette tip followed by 10 cycles through a 200-µl tip. 400,000 cells/well of dissociated cortical neurons were seeded in DMEM (Invitrogen) supplemented with 10% heat-inactivated horse serum in six-well plates on 12-mm glass coverslips precoated overnight with 50 μg/ml poly-d-lysine (Thermo Fisher Scientific; A3890401) at 37°C. Twenty hours after seeding, medium was changed to culture medium (neurobasal [Invitrogen], B27 supplement 2%, sodium pyruvate 1 mM, l-glutamine 2 mM, streptomycin 10 µg/ml, penicillin 10 U/ml), and 4 d after plating, cytosine arabinoside (AraC; 5 µM) was added. Neurons were fed every 4 d with 500 µl of fresh culture medium containing AraC and fixed for immunofluorescence microscopy after 8 d of culture.
For immunofluorescence microscopy of copper-dependent ATP7A trafficking, MDCK or mCCD culture medium containing 315 µM CuCl2 (Sigma-Aldrich; C3279), diluted from aqueous 1500× stock solution, was added to the cells (Greenough et al., 2004) overnight before fixation in the case of cysts, 4 or 5 h before fixation for MDCK or mCCD cells grown on Transwells, respectively. Basal copper conditions correspond to regular MDCK or mCCD culture medium, whose main source of copper is fetal bovine serum (10–20%) and copper concentration is less than 1 µM (Arredondo et al., 2000; Bauerly et al., 2004; Keenan et al., 2018). For copper washout, cells were treated with CuCl2 as described, washed once with culture medium, and then incubated for 4 h at 37°C in culture medium containing 200 µM copper chelating agent BCS (Santa Cruz Biotechnology; Sc-217698) and 50 µg/ml cycloheximide (protein synthesis inhibitor), with replacement with fresh medium every 80 min (Holloway et al., 2013).
To disrupt the microtubule network, cells were treated with 10 µM nocodazole (stock solution at 5 mg/ml in dimethyl sulfoxide [DMSO; Sigma-Aldrich; SML1665]) for 2 h at 37°C before immunofluorescence microscopy analysis. DMSO (maximal final concentration of 0.1%) treatment was used as negative control.
Genome engineering
PLEKHA7 KO and PDZD11 KO mCCD and Hap1 cells were described previously (Popov et al., 2015; Guerrera et al., 2016; Shah et al., 2018). CRISPR/Cas9 gene editing technology was used to generate PLEKHA5 and PLEKHA6 KO cells in wild-type (WT) or PLEKHA7 KO background to obtain single or double KO cell lines, respectively. The Genscript guide RNA (gRNA) designing tool (https://www.genscript.com/gRNA-design-tool.html) was used to determine the CRISPR target sequences (Table 1). After cloning the gRNAs into the BbsI site of Cas9 and the GFP-expressing px458 CRISPR plasmid (Addgene; catalogue no. 48138), specific background cells (Table 1) were transfected using Lipofectamine2000 (Invitrogen) and GFP-positive single-cell sorted as described previously (Guerrera et al., 2016; Shah et al., 2018). Single clones were further amplified and screened for KO using immunoblot and immunofluorescence microscopy analysis, before further validation by genotyping. Genomic DNA was purified using the DNeasy Blood and Tissue kit from QIAGEN (#69504), and the genomic locus surrounding the target region was amplified by PCR using specific primers (Table 1). Purified PCR products of Hap1 genotyping were subjected to Sanger sequencing (Microsynth, Switzerland), while products from mCCD and MDCK lines were first subcloned into the EcoRI-HindIII site of pcDNA3.1(+)/myc-His to separate alleles before being sequenced. Owing to missing regions in the genomic sequence of dog PLEKHA5, MDCK PLEKHA5 KO could not be genotyped.
TABLE 1:
Genome engineering of PLEKHA5 and PLEKHA6 in mCCD, Hap1, and MDCK cells.
| Cell line | Species | CRISPR target(s) | Cell background | Genotyping primers |
|---|---|---|---|---|
| mCCD PLEKHA6 KO | Mouse | GGTTCATAGAGCTTTTGCGC | mCCD N64 WT | gaggaattcCCAAGTTACCCCGAGAAGGG |
| GCCAGTCTTTTATGACGAGC | gagaagcttGGGAGGAGAGGACGTACCAT | |||
| mCCD PLEKHA6/7 KO | Mouse | GGTTCATAGAGCTTTTGCGC | mCCD N64 PLEKHA7 KO ( Guerrera et al., 2016; Shah et al., 2016, 2018) | gaggaattcCCAAGTTACCCCGAGAAGGG |
| GCCAGTCTTTTATGACGAGC | gagaagcttGGGAGGAGAGGACGTACCAT | |||
| Hap1 PLEKHA5 KO | Human | AATGCACCGGTTGTCAGACG | Hap1 WT | CAGGTAGGACAAAATACTGCCAC |
| CTGAAACCTAGCTGCAAACTGG | ||||
| Hap1 PLEKHA5/7 KO | Human | AATGCACCGGTTGTCAGACG | Hap1 PLEKHA7 KO ( Popov et al., 2015; Shah et al., 2018) | CAGGTAGGACAAAATACTGCCAC |
| CTGAAACCTAGCTGCAAACTGG | ||||
| MDCK PLEKHA5 KO | Dog | TGGACTTACGGGATCACCCG | MDCK II Tet-off WT | N/A |
| MDCK PLEKHA6 KO | Dog | CCACCCGAATGTTGATGAGC | MDCK II Tet-off WT | gaggaattcAAGCTGCTGGGCAAATTGTGTGA |
| gagaagcttCCCAGAACTACCGTCCAAGCAGCC |
Antibodies
The primary antibodies targeting the following proteins, raised in the detailed host species, were used at the indicated dilution for immunoblotting (IB), immunofluorescence microscopy (IF), or immunohistochemistry (IHC): rabbit PDZD11 (Rb29958; in-house [ Guerrera et al., 2016]; IB: 1/1000, IF, IHC: 1/100); rabbit PLEKHA7 (Rb30388; in-house [ Pulimeno et al., 2010]; IB: 1/5000, IF: 1/1000, IHC: 1/500); guinea pig PLEKHA7 (GP2737; in-house [ Guerrera et al., 2016]; IF: 1/500, IHC: 1/4000 or 1/300 for brain sections); mouse β-tubulin (32-2600; Thermo Fisher Scientific; IB: 1/3500); mouse α-tubulin (32-2500; Thermo Fisher Scientific; IF: 1/250); guinea pig α-tubulin (AA345 scFv-F2C; in house [ Guerreiro and Meraldi, 2019]; IF: 1/500); rabbit GFP (A-11122; Thermo Fisher Scientific; IB: 1/2000, IF: 1/200); mouse GFP (11814460001; Roche; IF: 1/100); mouse HA (32-6700; Thermo Fisher Scientific; IF: 1/150, IB: 1/1000); rabbit HA (sc-805; Santa Cruz; IF: 1/100); mouse myc (9E10; in-house; IB: 1/2); mouse E-cadherin (610181; BD Biosciences; IB: 1/5000, IF, IHC: 1/2500); mouse ZO-1 (33-9100; Thermo Fisher Scientific; IF: 1/1000); rat ZO-1 (R40.76; a kind gift from Daniel Goodenough [Harvard Medical School]); IF: 1/100); rabbit afadin (A0224; Sigma-Aldrich; IB: 1/8000, IF: 1/200); rabbit paracingulin (20893; in-house [ Pulimeno et al., 2011]; IB: 1/10 000, IF: 1/500); rabbit paracingulin (n.821; in-house [ Guillemot et al., 2008]; IF: 1/100); rabbit cingulin (C532; in-house [ Cardellini et al., 1996]; IF: 1/10,000, IB: 1/2000); mouse p120-catenin (8D11; a kind gift from A. Reynolds [Vanderbilt University] [ Wu et al., 1998]; IB: 1/2500, IF: 1/100); rabbit α-catenin (C2081; Sigma; IB: 1/8000); rabbit ß-catenin (C2206; Sigma; IB: 1/3500, IF: 1/500); rat nectin-3 (D084-3; MBL; IF: 1/100); rabbit ADAM10 (AB19026; Merck Millipore; IF: 1/300); mouse GP135 (3F2/D8; DSHB; IF: 1/5); mouse actin (MAB1501R; Merck Millipore; IB: 1/5000); mouse GFAP (G3893; Sigma-Aldrich; IF: 1/800); chicken GFAP (01-670-261; Invitrogen; IHC: 1/2000); mouse tubulin ß-III (801201; BioLegend; IF: 1/500); mouse ATP7A (sc-376467; Santa Cruz; IHC: 1/500); Armenian hamster PECAM-1 (MAB1398Z; Merck Millipore; IHC: 1/500); goat VE-cadherin (sc-6458; Santa Cruz; IF: 1/1000); mouse golgin-97 (A-21270 [CDF4]; Thermo Fisher Scientific; IF: 1/100); and mouse EEA1 (610457; BD Biosciences; IF: 1/75). The rabbit polyclonal antibody targeting ATP7A (RbCT78; IF, IHC: 1/500) (Steveson et al., 2003) was a kind gift from Betty Eipper (University of Connecticut Health Center). The rat polyclonal antibodies targeting either PLEKHA5 (RtSZR129; IB: 1/1000, IF, IHC: 1/100) or PLEKHA6 (RtSZR127; IB: 1/1000, IF, IHC: 1/100) were generated by immunization of rats (Polyclonal Antibody Production, Eurogentec) with purified N-terminally GST-fused C-terminal fragments of human PLEKHA5 (NP_061885; aa 817–1116) or PLEKHA6 (XP_011507599, aa 971–1297) produced in BL21DE3 bacteria.
Secondary antibodies for IF, from Jackson ImmunoResearch and diluted at 1/300, were anti-mouse (715-546-151), anti-rabbit (711-545-152), anti-rat (712-546-153), and anti–guinea pig (706-546-148) Alexa Fluor 488; anti-mouse (715-165-151), anti-rabbit (711-165-152), and anti-rat (712-166-153) Cy3; anti–guinea pig (706-605-148), anti-rabbit (711-605-152) and anti-goat (705-606-147) Alexa Fluor 647; anti-rat (712-175-153) and anti-mouse (715-605-151) Cy5. Additionally, for mouse brain IHC, secondary antibodies, diluted at 1/250, were anti-Armenian hamster DyLightTM405 (127-475-160; Jackson ImmunoResearch); anti-mouse Alexa Fluor Plus 555 (A32727; Thermo Fisher Scientific); and anti-chicken Alexa Fluor Plus 488 (A32931; Thermo Fisher Scientific). Anti-mouse, anti-rabbit (1/20,000; Promega; W4021 and W4011, respectively) and anti-rat (1/10,000; Thermo Fisher Scientific; 62-9520). Horseradish peroxidase (HRP)-conjugated secondary antibodies were used for IB.
Plasmids
Constructs that have been described previously include CFP-HA, full-length human PDZD11 (GFP-, GST-, or -HA), and GST-fused WW (1–162) PLEKHA7 (Guerrera et al., 2016), GFP- and myc-tagged full-length (1–1121) and N-terminal region (1–562) of human PLEKHA7 (Paschoud et al., 2014), EGFP-tagged Tetraspanin-15 and Tetraspanin-33 (Shah et al., 2018), and GST-fused N-terminal (1–40) and ∆24 human PDZD11 (Rouaud et al., 2020). GFP-tagged WT (1462–1500) and ∆PDZ-binding (1462–1496) C-terminal regions of human ATP7A (NP_000043.4) were obtained by annealing of oligonucleotides and insertion in the pcDNA3.1(–) plasmid previously modified to contain N-terminal GFP (pcDNA3.1-GFP) (Paschoud et al., 2011). Full-length sequences of human PLEKHA5 (NM_019012) and PLEKHA6 (XM_011509297) isoforms identified by Y2H screen were synthesized by Genscript (https://www.genscript.com/gene_synthesis.html) and amplified by PCR with appropriate oligonucleotides for subsequent cloning. GST fusion of truncated sequences (PLEKHA5: WW [1–120], Cter [817–1116]; PLEKHA6: WW [1–116], Cter [971–1297]) were cloned into pGEX4T1 (EcoRI/BamHI-NotI for PLEKHA5; EcoRI-NotI for PLEKHA6) for isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible bacterial expression. For mammalian expression, full-length GFP-tagged PLEKHA5 (1–1116) and PLEKHA6 (1–1297) constructs were obtained by PCR and subcloned into the pcDNA3.1(–) plasmid (NotI-KpnI for PLEKHA5, NotI-HindIII for PLEKHA6) previously modified to contain N-terminal GFP (pcDNA3.1-GFP) (Paschoud et al., 2011). N-terminally HA-tagged full-length PLEKHA5 and PLEKHA6 and N-terminal PLEKHA7 (1–562) were amplified by PCR using HA-containing forward primer before cloning into pcDNA3.1(–) (NotI-KpnI for PLEKHA5, NotI-HindIII for PLEKHA6 and PLEKHA7). Full-length PLEKHA5 and PLEKHA6, tagged with GFP in N-terminal and myc at the C-terminus, were made by PCR and subcloning into the NotI-ClaI site of a pTre2Hyg plasmid already containing GFP-myc (Paschoud et al., 2014). All constructs were validated by sequencing (Microsynth, Switzerland).
Yeast two-hybrid screen
The Y2H screen was carried out by Hybrigenics (France) with ULTImate Y2H screening technology. The full-length sequence of human PDZD11 (NP_001357103.1) was cloned in the pB27 vector with N-terminal LexA, and this construct was used to screen a human placenta library (RP6) in the presence of 20 mM 3-amino-1,2,4-triazole.
To perform bioinformatic analysis, the analyzed protein sequences corresponding to the human PLEKHA7 (NM_175058.4), PLEKHA5 (NM_019012.5), and PLEKHA6 (XM_011509297.2) were aligned using T-Coffee (version 8.93) from EMBL-EBI. WW and PH domains are SMART domains proposed by databases; coiled-coil domains were detected using NCoils (version 1) from Expasy; proline-rich domains were obtained by a scan in the protein profile database PROSITE from Expasy. The two WW domains and PH domain of the three proteins were submitted to WebLogo 3.7.4 (Crooks et al., 2004) for graphical representation of the amino acid multiple sequence alignment of these regions.
Cell transfection and immunofluorescence microscopy
For immunofluorescence microscopy (IF) staining, cells were seeded either on 6.5-mm/0.4-µm-pore polyester 24-well tissue culture inserts (Transwell filters; Corning Costar; #3470) or on 12-mm glass coverslips in 24-well plates. For Hap1 cells, coverslips were precoated with 0.01% poly-l-lysine (Sigma-Aldrich; P4707) for 30 min at 37°C before plating. To study the localization of tagged proteins that are exogenously expressed, cells at 60%–80% confluence were transfected 1 d after seeding, using either Lipofectamine2000 (Invitrogen) or jetOPTIMUS (Polyplus) following the manufacturer’s guidelines, and processed for IF 48–72 h later. Cells on coverslips were washed twice with room temperature (RT) phosphate-buffered saline (PBS) before methanol (precold at –80°C) fixation for 8 min at –20°C. After three PBS washes, cells were permeabilized 5 min in PBS/Triton X-100 0.3% and blocked 20 min in blocking buffer (PBS/gelatin 0.2%/bovine serum albumin [BSA] 1%/Triton X-100 0.03%) before incubation with primary antibodies and diluted in blocking buffer, for either 1 h at RT or 16 h at 4°C. Following three washes with PBS/Triton X-100 (0.3%) and 15 min of blocking, secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI) (1µg/ml), diluted in blocking buffer, were applied for 30 min at 37°C, before final washes with PBS/Triton X-100 (0.3%) (three times) and PBS and mounting with Fluoromount-G (Invitrogen). An alternative IF protocol, used when staining endogenous PLEKHA5 and PLEKHA6, consists of, after methanol fixation, washing the cells three times with PBS before a 30-min blocking step in PBS/donkey serum 1% and incubation overnight with primary antibodies diluted in serum incubation buffer (PBS/BSA 1%/donkey serum 1%/Triton X-100 0.3%) at 4°C. After three washes in PBS (15 min each), cells were incubated in secondary antibodies and DAPI (1 µg/ml), diluted in serum incubation buffer, for 30 min at 37°C, before final washes with PBS (three times, 15 min each) and mounting with Fluoromount-G (Invitrogen).
Cells grown on Transwells were fixed by 16 h incubation in methanol at –20°C, followed by a 1-min treatment with acetone precooled at –20°C. Filters were excised manually using a razor blade and hydrated in IMF buffer (0.1% Triton X-100, 0.15 M NaCl, 5 mM EDTA, 20 mM HEPES, pH 7.5, 0.02% NaN3 as preservative) for 15 min at RT. After two washes with IMF, cells were blocked with IMF/donkey serum 1% for 30 min and then incubated with primary antibodies diluted in serum incubation IMF (IMF/BSA 1%/donkey serum 1%/Triton X-100 0.3%) overnight at 4°C. Three IMF washes (15 min each) were done before incubation with secondary antibodies (diluted in serum incubation IMF) for 2 h at RT and final washes with IMF (four times, 15 min each). For staining of golgin-97 and EEA1, cells were fixed for 20 min at RT in 4% paraformaldehyde before quenching with NH4Cl 5 mM (in PBS) for 15 min at RT. After excision of filters, cells were permeabilized 5 min in PBS/Triton X-100 (0.3%) and blocked 30 min in blocking buffer (PBS/BSA 1%/Triton X-100 [0.03%]) before incubation with primary antibodies, diluted in blocking buffer, for 16 h at 4°C. Following three washes with PBS (15 min each, under slight shaking), secondary antibodies, diluted in blocking buffer, were applied for 2 h at RT, before four final washes with PBS (15 min each, under slight shaking). The filters were placed on glass slides, cells facing up, and were mounted with Vectashield containing DAPI (VECTOR Laboratories) and covered by a glass coverslip.
Cysts were fixed with methanol and acetone mixed 1:1 for 11 min at –20°C before permeabilization with PBS containing 0.5% Triton X-100 (10 min at RT). For staining of golgin-97 and EEA1, cells were fixed for 20 min at RT in 3% paraformaldehyde before quenching with NH4Cl 5 mM (in PBS) for 15 min at RT and permeabilization for 5 min in PBS/Triton X-100 (0.5%). Immunostaining was then performed as described previously (Spadaro et al., 2017).
Slides were imaged on a Zeiss LSM800 confocal microscope using a 63×/1.4 NA oil immersion objective. Staining of nuclei with DAPI is colored in blue. Unless otherwise stated, scale bars correspond to 20 µm.
Mouse tissue immunohistochemistry
Wild-type C57BL/6J mice were obtained from in-house breeding colonies. Mice were group housed on a 12:12 h light–dark cycle at 22°C with free access to food and water. All animal studies were approved by and performed according to the guidelines of the Animal Care and Use Committee of the University of Geneva (under authorization no. GE133/20) and of the University of California, Berkeley (under AUP-2019-04-12038). To harvest epithelial tissues, mice were killed and tissues included in OCT medium and snap-frozen in liquid nitrogen-cooled isopentane. Frozen sections (5 µm) were air-dried, fixed with acetone at –20°C for 20 min, and rehydrated in PBS. After 30 min of blocking in PBS/donkey serum 1%, sections were incubated with primary antibodies (overnight at 4°C) and secondary antibodies (1 h at RT) diluted in PBS/BSA 1%/donkey serum 1%/Triton X-100 0.3%, each followed by three washes in PBS, and were finally mounted with Vectashield containing DAPI (VECTOR Laboratories) and covered by a glass coverslip. Sections were imaged on a Zeiss LSM800 confocal microscope using a 40×/1.3 NA oil immersion objective. For brain IHC, mice were killed and immediately perfused with PBS and 4% formaldehyde in PBS. Brains were postfixed in 4% paraformaldehyde for 24–48 h and stored in 30% sucrose in PBS solution for 48 h for cryoprotection. Brains were embedded and mounted in Tissue-Tek OCT compound (Sakura finetek), and 20 µm sections were cut using a cryostat (Leica). Brain sections were rehydrated in PBS, permeabilized using PBST (0.3% Triton X-100 in PBS) for 30 min, and incubated with blocking solution (5% normal goat serum or normal donkey serum in PBST) for 1 h followed by primary antibody incubation overnight at 4°C. After washing in PBS, sections were incubated in corresponding fluorescently conjugated secondary antibodies (1/250) for 2 h at RT. After washing in PBS, sections were mounted with VECTASHIELD Antifade Mounting Medium (Vector Laboratories; H-1000). Fluorescence images were taken with a confocal microscope (LSM880 Confocal; Zeiss).
Imaging quantifications
To quantify ATP7A localization between cytoplasmic/TGN and membrane-associated fractions (XY view), the integrated density of ATP7A signal in the cytoplasmic region (drawn inside the E-cadherin labeling with the polygon selection tool of FIJI) was divided by the integrated density of the signal in the entire cell area, that is, comprising also the membrane-associated ATP7A staining, using E-cadherin to define this region with the polygon selection tool of FIJI. Quantification of ATP7A distribution at cell–cell contacts (XZ view) after copper treatment was done by calculating the zonular percentage of ATP7A signal, which was obtained by dividing the integrated density of the signal in the zonular region, using ZO-1 to delimit the area (polygon selection tool of FIJI), by the integrated density of the signal at the entire cell–cell contact area using E-cadherin to determine this region. Colocalization between ATP7A and golgin-97 was quantified by Pearson’s correlation coefficients. Coefficients were determined using the Colocalization Threshold plug-in in FIJI software, applying autothresholding from the Costes method (images with “Pearson’s below threshold” superior to 0.1 were not considered). Two to five images from three different stainings were used, each image being used as replicate.
Cell and tissue lysates; immunoblot analysis
Cell lysates were obtained in 500 µl of RIPA buffer (NaCl 150 mM/Tris-HCl 40 mM, pH 7.5/ EDTA 2 mM/glycerol 10%/Triton X-100 1%/sodium deoxycholate 0.5%/SDS 0.2%) supplemented with protease inhibitor cocktail (Thermo Fisher Scientific; A32965) from 10-cm dishes, followed by sonication (8 s at 66% amplitude with a Branson sonifier). Solubilized proteins were clarified by centrifugation (15 min at 4°C, 13,000 rpm). Organ lysates were obtained by homogenization of the sample in 500 µl of lysis buffer A (Guillemot et al., 2012) using plastic micropestles. After 15 min of incubation on ice, samples were sonicated five times for 5 s at 66% (Branson sonifier) before clarification by centrifugation (40 min at 4°C, 13,000 rpm).
Samples were mixed with SDS loading buffer and boiled 5 min at 95°C before SDS–PAGE separation at 4°C. Proteins were transferred onto nitrocellulose (0.45 µm) membrane (100 V for 80 min or 70 V for 180 min, at 4°C), and blots were blocked in Tris-buffered saline (TBS)/Tween-20 0.1%/low-fat milk 20% for 1 h before incubation with primary antibody (diluted in TBS/Tween-20 0.1%/low-fat milk 10%) followed by secondary HRP-labeled antibody (same buffer) for 1 h and finally chemiluminescence (ECL) revelation, which was detected using Odyssey Imager (LI-COR). Numbers on the left of immunoblots correspond to sizes in kilodaltons.
Basolateral surface biotinylation
ATP7A levels along the basolateral surface were assessed by cell surface biotinylation (Greenough et al., 2004; Pase et al., 2004). Cells (400,000) were grown on 24-mm Transwells (Corning Costar; #3450) for 7 d. For copper stimulation, culture medium containing 315 µM CuCl2 was added to the apical and basal chambers and incubated for 4–5 h at 37°C. Biotin labeling and processing was performed at 4°C for 30 min, using the Pierce Cell Surface Protein Isolation Kit (Thermo Fisher Scientific; 89881). Sulfo-NHS-SS-Biotin was dissolved in PBS supplemented with MgCl2 (0.5 mM) and CaCl2 (1 mM) (PBS++) and placed in the basal chamber, and PBS++ containing 315 µM CuCl2 was placed in the apical chamber. Biotin was quenched for 10 min, and cells were scraped and transferred to a tube. After two washes with TBS (resuspension, centrifugation for 3 min at 500 × g, and removal of supernatant), cells were lysed in lysis buffer (NaCl 150 mM/Tris-HCl 50 mM, pH 7.5/Triton X-100 1%/EDTA 5 mM) containing protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific; A32959) and sonicated two times for 4 s at 45% (Branson sonifier). Lysates were incubated 30 min on ice, vortexing every 5 min for 5 s, before centrifugation at 13,000 rpm for 15 min and transfer of the clarified supernatant to new tubes. Biotinylated proteins were purified by overnight incubation with NeutrAvidin Agarose slurry from the kit. After incubation, beads were washed three times with lysis buffer, twice with high-salt buffer (NaCl 500 mM/ Tris-HCl 50 mM, pH 7.5), and once with no-salt buffer (Tris-HCl 10 mM, pH 7.5) before elution with sample buffer supplemented with dithiothreitol (DTT) (200 mM) and urea (250 mg/ml) for 30 min at RT, vortexing every 5 min, and 15 min at 37°C. Samples were then analyzed by immunoblotting, along with inputs prepared in sample buffer containing DTT and urea (15 min of incubation at 37°C). Quantification of the ATP7A chemiluminescence signal was carried out in Image Studio Lite (LI-COR). For each genotype, the signal of biotinylated ATP7A was normalized to the input signal and calculated relative to the WT.
Recombinant protein expression and GST pull downs
For the production of GST-fused proteins, Escherichia coli (BL21-DE3) cell swere transformed by heat shock with pGEX4T1 constructs, and expression was induced with 0.1 mM IPTG for 2 h at 37°C. Bacterial pellets were snap-frozen in liquid nitrogen before lysis in lysis buffer (PBS/Triton X-100 1%) supplemented with protease inhibitor cocktail (Thermo Fisher Scientific; A32965) and sonication five times at 55% (Branson sonifier). Cell debris were removed by centrifugation (13,000 rpm) for 15 min at 4°C, and GST-tagged baits contained in supernatants were normalized using Pierce Glutathione Magnetic Agarose Beads (Thermo Fisher Scientific; #78602) according to the manufacturer’s protocol, followed by Coomassie staining of SDS–PAGE. Prey and additional (for trimolecular pull downs) proteins were expressed in HEK293T cells (2,300,000 cells in a 10-cm dish) transfected with 10 µg of DNA using polyethylenimine (Polysciences; #23966-2) and 48 h later, after washing with PBS, lysing in CoIP buffer (NaCl 150 mM/Tris-HCl 20 mM, pH 7.5/Nonidet P-40 1%/EDTA 1 mM) supplemented with protease inhibitor cocktail (Thermo Fisher Scientific; A32965), applying sonication (8 s at 66%; Branson sonifier) and centrifugation (15 min at 13,000 rpm, at 4°C). Prey and additional protein loadings were normalized by immunoblotting.
For GST pull downs, 5 µg of GST-fused bait was coupled to Pierce Glutathione Magnetic Agarose Beads (Thermo Fisher Scientific; #78602), previously washed twice with equilibration buffer (Tris-HCl 125 mM, pH 7.4/NaCl 150 mM/DTT 1 mM/EDTA 1 mM), for 1 h 30 min at RT. Following incubation and three washes with PBS/milk 2%/Nonidet P-40 1%, beads were incubated at 4°C overnight or for 2 h with normalized preys HEK293T lysates. After three washes with CoIP buffer, proteins bound to the beads were eluated with 20 μl of SDS loading buffer boiled at 95°C for 5 min, before analysis by immunoblotting. Because lysates, and not purified proteins, were used as preys, it cannot be formally excluded that contaminating proteins from the HEK293T lysates may affect the results; however, it is unlikely that they are present in sufficiently high concentrations to affect results, as preys were significantly overexpressed.
Quantification of WT C-terminal ATP7A chemiluminescence signal intensity in GST-PDZD11 pull downs in the presence of WW-PLEKHAs was carried out in Image Studio Lite (LI-COR), normalized to bait signal (Ponceau S straining) and calculated relative to control additional protein (CFP-HA) value.
Immunoprecipitation
Immunoprecipitation of proteins was carried out as described previously (Guerrera et al., 2016). Lysates were obtained from 10-cm dishes by rinsing cells with PBS and incubating them in 500 µl of CoIP buffer (NaCl 150 mM/Tris-HCl 20 mM, pH 7.5/Nonidet P-40 1%/EDTA 1 mM) supplemented with protease inhibitor cocktail (Thermo Fisher Scientific; A32965) for 10 min at 4°C. After sonication (8 s at 66%; Branson sonifier) and centrifugation (15 min at 13,000 rpm, at 4°C), the supernatant was collected (cytoskeleton-soluble fraction). The pellet was resuspended in 50 µl of SDS buffer (SDS 1%/Tris-HCl 10 mM, pH 7.5/EDTA 2 mM/DTT 0.5 mM/PMSF 0.5 mM), sonicated 3 s at a power of 15% (Branson sonifier), incubated 5 min at 95°C, and clarified by centrifugation. Supernatant was brought to a volume of 500 µl with CoIP buffer and mixed with the cytoskeleton-soluble fraction to obtain the total cell lysate. Dynabeads protein G (20 µl) (or protein A for guinea pig anti-PLEKHA7) (Invitrogen) were coupled to antibodies (diluted in PBS/BSA 5%/Nonidet P-40 1%; 2 µl of preimmune or immune serum for anti-PLEKHA7, 10 µl for anti-PLEKHA5 and -6) at 4°C for 90 min. After two washes with PBS/BSA 5%/Nonidet P-40 1%, beads were incubated overnight at 4°C with 100–120 µl of total cell lysate and then washed three times with CoIP buffer. Immunoprecipitates were eluted in 20 µl SDS loading buffer and boiled 5 min at 95°C before analysis by SDS–PAGE and immunoblotting.
Intracellular labile copper imaging
Cells were seeded on 35-mm glass-bottom fluorodishes (WPI; FD35-100) and incubated for 48 h until confluent. On the day of imaging, after 4 h (MDCK) or 5 h (mCCD) of incubation at 37°C (5% CO2) in fresh culture medium at basal or elevated (315 µM, CuCl2 mixed as aqueous solution) copper, cells were washed with Live Cell Imaging Solution (LCIS; Thermo Fisher Scientific; A14291DJ) before loading with either 1 µM CF4 (diluted in LCIS) or 1 µM Ctrl-CF4-S2 probe (Xiao et al., 2018) for 20 min at 37°C. Cells were rinsed twice with LCIS and imaged in LCIS at 37°C on a Zeiss LSM780 confocal microscope using a 40×/1.2 NA water immersion objective, exciting the probe at 536 nm (HeNe543 laser) and collecting emission between 545 and 700 nm. Seven to twenty-two fields were acquired for each fluorodish. In FIJI, the signal was adjusted to threshold to remove background areas and the mean intensity of the image (limited to threshold) was measured; the average of the 7–22 intensities was calculated for each fluorodish (replicate). GraphPad Prism 8 software was then used to normalize replicates to the mean of WT cells in basal copper conditions.
RNA isolation and quantitative PCR
Confluent mCCD cells seeded in 35-mm dishes were incubated for 5 h in basal or elevated copper (315 µM CuCl2 [Sigma-Aldrich; C3279], diluted from an aqueous 1500× stock solution) medium before mRNA extraction using the NucleoSpin RNA Purification Kit from Macherey-Nagel (740955.50) and cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad; 1708891), following the manufacturer’s instructions. Quantitative PCR (qPCR) analysis was performed on 20 ng of cDNA (each in triplicate) with SYBR Select Master Mix for CFX (Thermo Fisher Scientific [Life Technologies]; 4472942) for mouse metallothionein-I (Fw: GAATGGACCCCAACTGCTC; Rv: GCAGCAGCTCTTCTTGCAG) (Wunderlich et al., 2010) and GAPDH (internal standard, Fw: GTGCAGTGCCAGCCTCGTCC; Rv: CTCGGCCTTGACTGTGCCGT).
ICP-MS
Cells (600,000/well) were cultured in six-well plates for 72 h before incubation in basal or elevated copper (315 µM CuCl2 [Sigma-Aldrich; C3279], diluted from an aqueous 1500× stock solution) medium for 4 h. To measure the efflux of copper, copper-treated cells were washed once with culture medium and then incubated for 30 min in fresh culture medium (basal copper). For mineralization, cells were rinsed three times with ice-cold PBS and incubated 16 h at RT in trace-metals grade concentrated nitric acid (215 µl/well) (VWR [Normatom]; 83872.290). After incubation, a volume of 150 µl was taken and completed up to 3.3 ml with water in 15 ml metal-free tubes, ending to a final solvent composition of sample matrix of 4.5% HNO3 in water (vol/vol). The analyses were performed by the Mass Spectrometry Core Facility from the Faculty of Sciences of the University of Geneva, usinga 7700× inductively coupled plasma mass spectrometer (ICP-MS) equipped with a Micromist nebulizer and a Scott nebulizing chamber (Agilent Technologies). The autosampler was an ASX-500 series (Cetac), and the software used for data acquisition was MassHunter 4.2 (C.01.02 version). Copper content was determined by measuring 63Cu and 65Cu in helium (He) mode with the He flow set to 4.3 ml/min, in triplicate instrumental measurements. The internal standard was 20 ppb of rhodium, diluted from ICP standard monoelement solution of rhodium (SCP Science; 140-052-450). Sample measurements were compared with a calibration curve established with dilutions of ICP standard monoelement solution of copper (SCP Science; 140-051-290). Two rounds of experiments were conducted, each composed of three repetitions of every copper-treatment condition. GraphPad Prism 8 software was used to normalize all conditions to the mean of WT cells in elevated copper conditions (relative cellular copper content).
Cell viability analysis
Cells were seeded in a 96-well plate (4000 cells/well, each condition in triplicate) 72 h before copper treatment, to reach confluency. Fresh culture medium at basal or elevated copper (315 µM CuCl2 [Sigma-Aldrich; C3279], diluted from aqueous 1500× stock solution) was added for 20 h before fixation for 20 min at RT in 4% paraformaldehyde. After two washes with PBS, cells were stained for 20 min at RT with crystal violet (0.5%)/ethanol (30%)/water, washed three times with water, and then dried for 2 d at RT (Feoktistova et al., 2016). Colorant was dissolved in 10% acetic acid for 10 min, and absorbance at 590 was measured with Cytation3 plate reader. For each experiment, the triplicate for each condition was averaged and, after subtraction of the blank (empty wells), normalized to the value of the basal copper level condition of the corresponding genotype.
Statistical analysis
Statistical significance was determined using GraphPad Prism 8 software; sample size (n), p values, and statistical tests performed are specified in figure legends. Values are shown as mean ± SD. Significance has been determined as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. For multiple comparison tests, after analysis of variance (ANOVA) results showing significant difference, Dunnett’s test was used to compare every mean to control mean.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (31003A_172809) (to S. C.), by the State of Geneva, by the Service Egalité of the University of Geneva (Subside Tremplin to S.S.), and by National Institutes of Health Grant GM 79465 (to C.J.C.). We thank Arielle Flinois and Marine Laporte for protocols of cysts and neuronal culture, Betty Eipper and Scot Leary (University of Saskatchewan, Canada) for the gift of anti-ATP7A antibodies, Quentin Devos and Thomas Di Mattia for informatic help, and Sophie Michalet and Emmanuel Varesio for help with ICP-MS.
Abbreviations used:
- A427
human lung carcinoma cell line
- ADAM10
a disintegrin and metalloproteinase domain-containing protein 10
- AIPP1
ATPase-interacting PDZ protein
- AJ
adherens junction
- AP-1
adaptor protein 1
- AP-2
adaptor protein 2
- AraC
cytosine arabinoside
- ATP7A
Menkes copper ATPase
- ATP7B
Wilson copper ATPase
- BCS
bathocuproinedisulfonic acid
- bEnd.3
mouse brain microvascular endothelial (endothelioma) cell line
- BSA
bovine serum albumin
- Caco-2
human intestinal colon carcinoma cells
- CC domain
coiled-coil domain
- CF4 probe
copper Fluor-4 probe
- CFP
cyan fluorescent protein
- CGN
cingulin
- CGNL1
paracingulin
- ctn
catenin
- Ctrl-CF4-S2 probe
copper Fluor-4-Sulfur-2 probe
- Cu
copper
- DAPI
4 ′,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified eagle medium
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- E-cadherin
epithelial cadherin
- ECL
enhanced chemiluminescence
- EDTA
ethylenediaminetetraacetic acid
- EEA1
early endosome antigen 1
- EGF
enhanced green fluorescent protein
- EGF
epidermal growth factor
- Eph4
mouse mammary epithelial cells
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- GFP
green fluorescent protein
- GP135
podocalyxin
- gRNA
guide RNA
- GST
glutathione- S-transferase
- H5V
mouse heart endothelial cell line
- HA
hemagglutinin
- HaCaT
human keratinocyte cell line
- Hap1
haploid human cells
- HBSS
Hanks’ balanced salt solution
- HEK293T
human embryonic kidney epithelial cells
- HRP
horseradish peroxidase
- HUVEC
human umbilical vascular endothelial cells
- IB
immunoblot
- ICP-MS
inductively coupled plasma mass spectrometry
- IF
immunofluorescence
- Ig-like
immunoglobulin-like
- IHC
immunohistochemistry
- IPTG
isopropyl β- d-1-thiogalactopyranoside
- kDa
kilodaltons
- KO
knock-out
- LC
locus coeruleus
- LCIS
live cell imaging solution
- LOD
limit of detection
- mCCD
mouse cortical collecting duct cells
- MDCK
Madin-Darby canine kidney cells
- meEC
ciliated aortic mouse embryonic endothelial cells
- MTT-I
metallothionein-I
- NA
numerical aperture
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PDZ domain
Psd95-Dlg1-ZO-1 domain
- PDZD11
PDZ domain-containing protein 11
- PECAM-1
platelet endothelial cell adhesion molecule
- PEPP2
phosphatidylinositol-three-phosphate-binding PH-domain protein-2
- PEPP3
phosphatidylinositol-three-phosphate-binding PH-domain protein-3
- PH domain
pleckstrin homology domain
- PLEKHA
pleckstrin homology domain-containing family A
- PLEKHA5
pleckstrin homology domain-containing family A member 5
- PLEKHA6
pleckstrin homology domain-containing family A member 6
- PLEKHA7
pleckstrin homology domain-containing family A member 7
- PMCA
plasma membrane calcium ATPase
- PMSF
phenylmethylsulfonyl fluoride
- PPBM
phosphatidylinositol (3,4,5)P3-binding motif
- Pro-rich domain
proline-rich domain
- PtdIns
phosphatidylinositol
- qPCR
quantitative Polymerase Chain Reaction
- RT
room temperature
- SD
standard deviation
- SLC5A6
sodium-dependent multivitamin transporter
- SMEM
spinner minimum essential medium
- TBS
tris buffered saline
- TGN
trans-Golgi network
- Tspan15
tetraspanin-15
- Tspan33
tetraspanin-33
- VE-cadherin
vascular endothelial cadherin
- WT
wild-type
- WW domain
Trp-Trp domain
- Y2H screen
yeast two-hybrid screen
- YFP
yellow fluorescent protein
- ZO-1
zonula occludens-1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-07-0355) on October 6, 2021.
REFERENCES
- Ackerman CM, Chang CJ (2018). Copper signaling in the brain and beyond. J Biol Chem 293, 4628–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredondo M, Uauy R, Gonzalez M (2000). Regulation of copper uptake and transport in intestinal cell monolayers by acute and chronic copper exposure. Biochim Biophys Acta 1474, 169–176. [DOI] [PubMed] [Google Scholar]
- Bandmann O, Weiss KH, Kaler SG (2015). Wilson’s disease and other neurological copper disorders. Lancet Neurol 14, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbitoff YA, Serebryakova EA, Nasykhova YA, Predeus AV, Polev DE, Shuvalova AR, Vasiliev EV, Urazov SP, Sarana AM, Scherbak SG, et al. (2018). Identification of novel candidate markers of type 2 iabetes and obesity in Russia by exome sequencing with a limited sample size. Genes (Basel) 9, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerly KA, Kelleher SL, Lönnerdal B (2004). Functional and molecular responses of suckling rat pups and human intestinal Caco-2 cells to copper treatment. J Nutr Biochem 15, 155–162. [DOI] [PubMed] [Google Scholar]
- Bayes A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, Grant SG (2011). Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci 14, 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camakaris J, Petris MJ, Bailey L, Shen P, Lockhart P, Glover TW, Barcroft C, Patton J, Mercer JF (1995). Gene amplification of the Menkes (MNK; ATP7A) P-type ATPase gene of CHO cells is associated with copper resistance and enhanced copper efflux. Hum Mol Genet 4, 2117–2123. [DOI] [PubMed] [Google Scholar]
- Camakaris J, Voskoboinik I, Mercer JF (1999). Molecular mechanisms of copper homeostasis. Biochem Biophys Res Commun 261, 225–232. [DOI] [PubMed] [Google Scholar]
- Cardellini P, Davanzo G, Citi S (1996). Tight junctions in early amphibian development: detection of junctional cingulin from the 2-cell stage and its localization at the boundary of distinct membrane domains in dividing blastomeres in low calcium. Dev Dyn 207, 104–113. [DOI] [PubMed] [Google Scholar]
- Chang CJ (2015). Searching for harmony in transition-metal signaling. Nat Chem Biol 11, 744–747. [DOI] [PubMed] [Google Scholar]
- Chassefeyre R, Martinez-Hernandez J, Bertaso F, Bouquier N, Blot B, Laporte M, Fraboulet S, Coute Y, Devoy A, Isaacs AM, et al. (2015). Regulation of postsynaptic function by the dementia-related ESCRT-III subunit CHMP2B. J Neurosci 35, 3155–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicka MC, Strehler EE (2003). Alternative splicing of the first intracellular loop of plasma membrane Ca2+-ATPase isoform 2 alters its membrane targeting. J Biol Chem 278, 18464–18470. [DOI] [PubMed] [Google Scholar]
- Choi BS, Zheng W (2009). Copper transport to the brain by the blood-brain barrier and blood-CSF barrier. Brain Res 1248, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold C, Coventry J, Ponnambalam S, Monaco AP (2004). Actin and microtubule regulation of trans-Golgi network architecture, and copper-dependent protein transport to the cell surface. Mol Membr Biol 21, 59–66. [DOI] [PubMed] [Google Scholar]
- Cobbold C, Ponnambalam S, Francis MJ, Monaco AP (2002). Novel membrane traffic steps regulate the exocytosis of the Menkes disease ATPase. Hum Mol Genet 11, 2855–2866. [DOI] [PubMed] [Google Scholar]
- Cox LL, Cox TC, Moreno Uribe LM, Zhu Y, Richter CT, Nidey N, Standley JM, Deng M, Blue E, Chong JX, et al. (2018). Mutations in the epithelial cadherin-p120-catenin omplex cause Mendelian non-syndromic cleft lip with or without cleft palate. Am J Hum Genet 102, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czlonkowska A, Litwin T, Dusek P, Ferenci P, Lutsenko S, Medici V, Rybakowski JK, Weiss KH, Schilsky ML (2018). Wilson disease. Nat Rev Dis Primers 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulagala AC, Bridges MC, Kourtidis A (2019). E-cadherin beyond structure: a signaling hub in colon homeostasis and disease. Int J Mol Sci 20, 2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268. [DOI] [PubMed] [Google Scholar]
- Dierick HA, Adam AN, Escara-Wilke JF, Glover TW (1997). Immunocytochemical localization of the Menkes copper transport protein (ATP7A) to the trans-Golgi network. Hum Mol Genet 6, 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR (2000). Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J 351, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Leverkus M (2016). Crystal violet assay for determining viability of cultured cells. Cold Spring Harb Protoc 2016, pdb prot087379. [DOI] [PubMed] [Google Scholar]
- Francis MJ, Jones EE, Levy ER, Ponnambalam S, Chelly J, Monaco AP (1998). A Golgi localization signal identified in the Menkes recombinant protein. Hum Mol Genet 7, 1245–1252. [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. (2014). De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goellner GM, DeMarco SJ, Strehler EE (2003). Characterization of PISP, a novel single-PDZ protein that binds to all plasma membrane Ca2+-ATPase b-splice variants. Ann NY Acad Sci 986, 461–471. [DOI] [PubMed] [Google Scholar]
- Goodyer ID, Jones EE, Monaco AP, Francis MJ (1999). Characterization of the Menkes protein copper-binding domains and their role in copper-induced protein relocalization. Hum Mol Genet 8, 1473–1478. [DOI] [PubMed] [Google Scholar]
- Greenough M, Pase L, Voskoboinik I, Petris MJ, O’Brien AW, Camakaris J (2004). Signals regulating trafficking of Menkes (MNK; ATP7A) copper-translocating P-type ATPase in polarized MDCK cells. Am J Physiol Cell Physiol 287, C1463–C1471. [DOI] [PubMed] [Google Scholar]
- Guerreiro A, Meraldi P (2019). AA344 and AA345 antibodies recognize the microtubule network in human cells by immunofluorescence. Antibody Rep, DOI: 10.24450/journals/abrep.2019.e17. [Google Scholar]
- Guerrera D, Shah J, Vasileva E, Sluysmans S, Mean I, Jond L, Poser I, Mann M, Hyman AA, Citi S (2016). PLEKHA7 recruits PDZD11 to adherens junctions to stabilize nectins. J Biol Chem 291, 11016–11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot L, Paschoud S, Jond L, Foglia A, Citi S (2008). Paracingulin regulates the activity of Rac1 and RhoA GTPases by recruiting Tiam1 and GEF-H1 to epithelial junctions. Mol Biol Cell 19, 4442–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot L, Schneider Y, Brun P, Castagliuolo I, Pizzuti D, Martines D, Jond L, Bongiovanni M, Citi S (2012). Cingulin is dispensable for epithelial barrier function and tight junction structure, and plays a role in the control of claudin-2 expression and response to duodenal mucosa injury. J Cell Sci 125, 5005–5012. [DOI] [PubMed] [Google Scholar]
- Hartwig C, Zlatic SA, Wallin M, Vrailas-Mortimer A, Fahrni CJ, Faundez V (2019). Trafficking mechanisms of P-type ATPase copper transporters. Curr Opin Cell Biol 59, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway ZG, Grabski R, Szul T, Styers ML, Coventry JA, Monaco AP, Sztul E (2007). Activation of ADP-ribosylation factor regulates biogenesis of the ATP7A-containing trans-Golgi network compartment and its Cu-induced trafficking. Am J Physiol Cell Physiol 293, C1753–C1767. [DOI] [PubMed] [Google Scholar]
- Holloway ZG, Velayos-Baeza A, Howell GJ, Levecque C, Ponnambalam S, Sztul E, Monaco AP (2013). Trafficking of the Menkes copper transporter ATP7A is regulated by clathrin-, AP-2-, AP-1-, and Rab22-dependent steps. Mol Biol Cell 24, 1735–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liao X, Luo J, Liu H, Zhong S, Chen J (2020). Expression of circular RNAs in the vascular dementia rats. Neurosci Lett 735, 135087. [DOI] [PubMed] [Google Scholar]
- Huster D, Finegold MJ, Morgan CT, Burkhead JL, Nixon R, Vanderwerf SM, Gilliam CT, Lutsenko S (2006). Consequences of copper accumulation in the livers of the Atp7b-/- (Wilson disease gene) knockout mice. Am J Pathol 168, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T, Nishimura M, Sugimura H, Igarashi H, Ozawa F, Shinmura K, Suzuki M, Tanaka M, Kino I (1996). Localization of Menkes gene expression in the mouse brain; its association with neurological manifestations in Menkes model mice. Acta Neuropathol 91, 482–488. [DOI] [PubMed] [Google Scholar]
- Jamain S, Cichon S, Etain B, Muhleisen TW, Georgi A, Zidane N, Chevallier L, Deshommes J, Nicolas A, Henrion A, et al. (2014). Common and rare variant analysis in early-onset bipolar disorder vulnerability. PLoS One 9, e104326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilaveanu LB, Parisi F, Barr ML, Zito CR, Cruz-Munoz W, Kerbel RS, Rimm DL, Bosenberg MW, Halaban R, Kluger Y, Kluger HM (2015). PLEKHA5 as a biomarker and potential mediator of melanoma brain metastasis. Clin Cancer Res 21, 2138–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaler SG (2011). ATP7A-related copper transport diseases-emerging concepts and future trends. Nat Rev Neurol 7, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan J, Horgan K, Clynes M, Sinkunaite I, Ward P, Murphy R, O’Sullivan F (2018). Unexpected fluctuations of trace element levels in cell culture medium in vitro: caveat emptor. In Vitro Cell Dev Biol Anim 54, 555–558. [DOI] [PubMed] [Google Scholar]
- Kodama H, Meguro Y, Abe T, Rayner MH, Suzuki KT, Kobayashi S, Nishimura M (1991). Genetic expression of Menkes disease in cultured astrocytes of the macular mouse. J Inherit Metab Dis 14, 896–901. [DOI] [PubMed] [Google Scholar]
- Kuo YM, Gitschier J, Packman S (1997). Developmental expression of the mouse mottled and toxic milk genes suggests distinct functions for the Menkes and Wilson disease copper transporters. Hum Mol Genet 6, 1043–1049. [DOI] [PubMed] [Google Scholar]
- La Fontaine S, Firth SD, Lockhart PJ, Brooks H, Camakaris J, Mercer JF (1999). Intracellular localization and loss of copper responsiveness of Mnk, the murine homologue of the Menkes protein, in cells from blotchy (Mo blo) and brindled (Mo br) mouse mutants. Hum Mol Genet 8, 1069–1075. [DOI] [PubMed] [Google Scholar]
- La Fontaine S, Firth SD, Lockhart PJ, Brooks H, Parton RG, Camakaris J, Mercer JF (1998). Functional analysis and intracellular localization of the human menkes protein (MNK) stably expressed from a cDNA construct in Chinese hamster ovary cells (CHO-K1). Hum Mol Genet 7, 1293–1300. [DOI] [PubMed] [Google Scholar]
- La Fontaine S, Mercer JF (2007). Trafficking of the copper-ATPases, ATP7A and ATP7B: role in copper homeostasis. Arch Biochem Biophys 463, 149–167. [DOI] [PubMed] [Google Scholar]
- Lee MC, Shei W, Chan AS, Chua BT, Goh SR, Chong YF, Hilmy MH, Nongpiur ME, Baskaran M, Khor CC, et al. (2017). Primary angle closure glaucoma (PACG) susceptibility gene PLEKHA7 encodes a novel Rac1/Cdc42 GAP that modulates cell migration and blood-aqueous barrier function. Hum Mol Genet 26, 4011–4027. [DOI] [PubMed] [Google Scholar]
- Linz R, Barnes NL, Zimnicka AM, Kaplan JH, Eipper B, Lutsenko S (2008). Intracellular targeting of copper-transporting ATPase ATP7A in a normal and Atp7b-/- kidney. Am J Physiol Renal Physiol 294, F53–F61. [DOI] [PubMed] [Google Scholar]
- Liu J, Adhav R, Miao K, Su SM, Mo L, Chan UI, Zhang X, Xu J, Li J, Shu X, et al. (2020). Characterization of BRCA1-deficient premalignant tissues and cancers identifies Plekha5 as a tumor metastasis suppressor. Nat Commun 11, 4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsenko S, Barnes NL, Bartee MY, Dmitriev OY (2007). Function and regulation of human copper-transporting ATPases. Physiol Rev 87, 1011–1046. [DOI] [PubMed] [Google Scholar]
- Lutsenko S, Petrukhin K, Cooper MJ, Gilliam CT, Kaplan JH (1997). N-terminal domains of human copper-transporting adenosine triphosphatases (the Wilson’s and Menkes disease proteins) bind copper selectively in vivo and in vitro with stoichiometry of one copper per metal-binding repeat. J Biol Chem 272, 18939–18944. [DOI] [PubMed] [Google Scholar]
- Lutsenko S, Washington-Hughes C, Ralle M, Schmidt K (2019). Copper and the brain noradrenergic system. J Biol Inorg Chem 24, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M (2008). Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135, 948–959. [DOI] [PubMed] [Google Scholar]
- Mercer JF, Lazdins I, Stevenson T, Camakaris J, Danks DM (1981). Copper induction of translatable metallothionein messenger RNA. Biosci Rep 1, 793–800. [DOI] [PubMed] [Google Scholar]
- Monty JF, Llanos RM, Mercer JF, Kramer DR (2005). Copper exposure induces trafficking of the menkes protein in intestinal epithelium of ATP7A transgenic mice. J Nutr 135, 2762–2766. [DOI] [PubMed] [Google Scholar]
- Nabokina SM, Subramanian VS, Said HM (2011). Association of PDZ-containing protein PDZD11 with the human sodium-dependent multivitamin transporter. Am J Physiol Gastrointest Liver Physiol 300, G561–G567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt T, Ohrvik H, Thiele DJ (2012). Charting the travels of copper in eukaryotes from yeast to mammals. Biochim Biophys Acta 1823, 1580–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyasae L, Bustos R, Braiterman L, Eipper B, Hubbard A (2007). Dynamics of endogenous ATP7A (Menkes protein) in intestinal epithelial cells: copper-dependent redistribution between two intracellular sites. Am J Physiol Gastrointest Liver Physiol 292, G1181–G1194. [DOI] [PubMed] [Google Scholar]
- Pandya NJ, Koopmans F, Slotman JA, Paliukhovich I, Houtsmuller AB, Smit AB, Li KW (2017). Correlation profiling of brain sub-cellular proteomes reveals co-assembly of synaptic proteins and subcellular distribution. Sci Rep 7, 12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschoud S, Jond L, Guerrera D, Citi S (2014). PLEKHA7 modulates epithelial tight junction barrier function. Tissue Barriers 2, e28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschoud S, Yu D, Pulimeno P, Jond L, Turner JR, Citi S (2011). Cingulin and paracingulin show similar dynamic behaviour, but are recruited independently to junctions. Mol Membrane Biol 28, 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pase L, Voskoboinik I, Greenough M, Camakaris J (2004). Copper stimulates trafficking of a distinct pool of the Menkes copper ATPase (ATP7A) to the plasma membrane and diverts it into a rapid recycling pool. Biochem J 378, 1031–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petris MJ, Mercer JF (1999). The Menkes protein (ATP7A; MNK) cycles via the plasma membrane both in basal and elevated extracellular copper using a C-terminal di-leucine endocytic signal. Hum Mol Genet 8, 2107–2115. [DOI] [PubMed] [Google Scholar]
- Petris MJ, Mercer JF, Culvenor JG, Lockhart P, Gleeson PA, Camakaris J (1996). Ligand-regulated transport of the Menkes copper P-type ATPase efflux pump from the Golgi apparatus to the plasma membrane: a novel mechanism of regulated trafficking. EMBO J 15, 6084–6095. [PMC free article] [PubMed] [Google Scholar]
- Petris MJ, Voskoboinik I, Cater M, Smith K, Kim BE, Llanos RM, Strausak D, Camakaris J, Mercer JF (2002). Copper-regulated trafficking of the Menkes disease copper ATPase is associated with formation of a phosphorylated catalytic intermediate. J Biol Chem 277, 46736–46742. [DOI] [PubMed] [Google Scholar]
- Polishchuk R, Lutsenko S (2013). Golgi in copper homeostasis: a view from the membrane trafficking field. Histochem Cell Biol 140, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov LM, Marceau CD, Starkl PM, Lumb JH, Shah J, Guerrera D, Cooper RL, Merakou C, Bouley DM, Meng W, et al. (2015). The adherens junctions control susceptibility to Staphylococcus aureus alpha-toxin. Proc Natl Acad Sci USA 112, 14337–14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulimeno P, Bauer C, Stutz J, Citi S (2010). PLEKHA7 is an adherens junction protein with a tissue distribution and subcellular localization distinct from ZO-1 and E-cadherin. PLoS One 5, 10.1371/journal.pone.0012207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulimeno P, Paschoud S, Citi S (2011). A role for ZO-1 and PLEKHA7 in recruiting paracingulin to tight and adherens junctions of epithelial cells. J Biol Chem 286, 16743–16750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Tiffany-Castiglioni E, Welsh J, Harris ED (1998). Copper efflux from murine microvascular cells requires expression of the menkes disease Cu-ATPase. J Nutr 128, 1276–1282. [DOI] [PubMed] [Google Scholar]
- Rouaud F, Tessaro F, Aimaretti L, Scapozza L, Citi S (2020). Cooperative binding of the tandem WW domains of PLEKHA7 to PDZD11 promotes conformation-dependent interaction with tetraspanin 33. J Biol Chem 295, 9299–9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaud F, Vasileva E, Spadaro D, Tsukita S, Citi S (2019). R40.76 binds to the alpha domain of ZO-1: role of ZO-1 (alpha+) in epithelial differentiation and mechano-sensing. Tissue Barriers 7, e1653748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilsky ML (1996). Wilson disease: genetic basis of copper toxicity and natural history. Semin Liver Dis 16, 83–95. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Ralle M, Schaffer T, Jayakanthan S, Bari B, Muchenditsi A, Lutsenko S (2018). ATP7A and ATP7B copper transporters have distinct functions in the regulation of neuronal dopamine-beta-hydroxylase. J Biol Chem 293, 20085–20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Guerrera D, Vasileva E, Sluysmans S, Bertels E, Citi S (2016). PLEKHA7: cytoskeletal adaptor protein at center stage in junctional organization and signaling. Int J Biochem Cell Biol 75, 112–116. [DOI] [PubMed] [Google Scholar]
- Shah J, Rouaud F, Guerrera D, Vasileva E, Popov LM, Kelley WL, Rubinstein E, Carette JE, Amieva MR, Citi S (2018). A dock-and-lock mechanism clusters ADAM10 at cell-cell junctions to promote alpha-toxin cytotoxicity. Cell Rep 25, 2132–2147.e2137. [DOI] [PubMed] [Google Scholar]
- Skjorringe T, Amstrup Pedersen P, Salling Thorborg S, Nissen P, Gourdon P, Birk Moller L (2017). Characterization of ATP7A missense mutants suggests a correlation between intracellular trafficking and severity of Menkes disease. Sci Rep 7, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluysmans S, Méan I, Jond L, Citi S (2021). WW, PH and C-terminal domains cooperate to direct the subcellular localizations of PLEKHA5, PLEKHA6 and PLEKHA7. Front Cell Dev Biol 9, 729444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro D, Le S, Laroche T, Mean I, Jond L, Yan J, Citi S (2017). Tension-dependent stretching activates ZO-1 to control the junctional localization of its interactors. Curr Biol 27, 3783–3795.e3788. [DOI] [PubMed] [Google Scholar]
- Spellmann I, Rujescu D, Musil R, Meyerwas S, Giegling I, Genius J, Zill P, Dehning S, Cerovecki A, Seemuller F, et al. (2014). Pleckstrin homology domain containing 6 protein (PLEKHA6) polymorphisms are associated with psychopathology and response to treatment in schizophrenic patients. Prog Neuropsychopharmacol Biol Psychiatry 51, 190–195. [DOI] [PubMed] [Google Scholar]
- Stephenson SE, Dubach D, Lim CM, Mercer JF, La Fontaine S (2005). A single PDZ domain protein interacts with the Menkes copper ATPase, ATP7A. A new protein implicated in copper homeostasis. J Biol Chem 280, 33270–33279. [DOI] [PubMed] [Google Scholar]
- Steveson TC, Ciccotosto GD, Ma XM, Mueller GP, Mains RE, Eipper BA (2003). Menkes protein contributes to the function of peptidylglycine alpha-amidating monooxygenase. Endocrinology 144, 188–200. [DOI] [PubMed] [Google Scholar]
- Strausak D, La Fontaine S, Hill J, Firth SD, Lockhart PJ, Mercer JF (1999). The role of GMXCXXC metal binding sites in the copper-induced redistribution of the Menkes protein. J Biol Chem 274, 11170–11177. [DOI] [PubMed] [Google Scholar]
- Subramanian VS, Marchant JS, Boulware MJ, Ma TY, Said HM (2009). Membrane targeting and intracellular trafficking of the human sodium-dependent multivitamin transporter in polarized epithelial cells. Am J Physiol Cell Physiol 296, C663–C671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavano S, Taverna E, Kalebic N, Haffner C, Namba T, Dahl A, Wilsch-Brauninger M, Paridaen J, Huttner WB (2018). Insm1 induces neural progenitor delamination in developing neocortex via downregulation of the adherens junction belt-specific protein Plekha7. Neuron 97, 1–16. [DOI] [PubMed] [Google Scholar]
- Telianidis J, Hung YH, Materia S, Fontaine SL (2013). Role of the P-type ATPases, ATP7A and ATP7B in brain copper homeostasis. Front Aging Neurosci 5, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa I, Fox HS, Bastola D (2015). Coexpression network analysis of miRNA-142 overexpression in neuronal cells. Biomed Res Int 2015, 921517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva E, Sluysmans S, Bochaton-Piallat ML, Citi S (2017). Cell-specific diversity in the expression and organization of cytoplasmic plaque proteins of apical junctions. Ann NY Acad Sci 1405, 160–166. [DOI] [PubMed] [Google Scholar]
- Veldhuis NA, Gaeth AP, Pearson RB, Gabriel K, Camakaris J (2009). The multi-layered regulation of copper translocating P-type ATPases. Biometals 22, 177–190. [DOI] [PubMed] [Google Scholar]
- Wu J, Mariner DJ, Thoreson MA, Reynolds AB (1998). Production and characterization of monoclonal antibodies to the catenin p120ctn. Hybridoma 17, 175–183. [DOI] [PubMed] [Google Scholar]
- Wunderlich KA, Leveillard T, Penkowa M, Zrenner E, Perez MT (2010). Altered expression of metallothionein-I and -II and their receptor megalin in inherited photoreceptor degeneration. Invest Ophthalmol Vis Sci 51, 4809–4820. [DOI] [PubMed] [Google Scholar]
- Wythe JD, Jurynec MJ, Urness LD, Jones CA, Sabeh MK, Werdich AA, Sato M, Yost HJ, Grunwald DJ, Macrae CA, Li DY (2011). Hadp1, a newly identified pleckstrin homology domain protein, is required for cardiac contractility in zebrafish. Dis Model Mech 4, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Ackerman CM, Carroll EC, Jia S, Hoagland A, Chan J, Thai B, Liu CS, Isacoff EY, Chang CJ (2018). Copper regulates rest-activity cycles through the locus coeruleus-norepinephrine system. Nat Chem Biol 14, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Heiny ME, Suzuki M, Gitlin JD (1996). Biochemical characterization and intracellular localization of the Menkes disease protein. Proc Natl Acad Sci USA 93, 14030–14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Kaler SG (2015). Direct interactions of adaptor protein complexes 1 and 2 with the copper transporter ATP7A mediate its anterograde and retrograde trafficking. Hum Mol Genet 24, 2411–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic S, Comstra HS, Gokhale A, Petris MJ, Faundez V (2015). Molecular basis of neurodegeneration and neurodevelopmental defects in Menkes disease. Neurobiol Dis 81, 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Cox TC (2013). A likely role for the PH-domain containing protein, PEPP2/PLEKHA5, at the membrane-microtubule cytoskeleton interface. BIOCELL 37, 55–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.