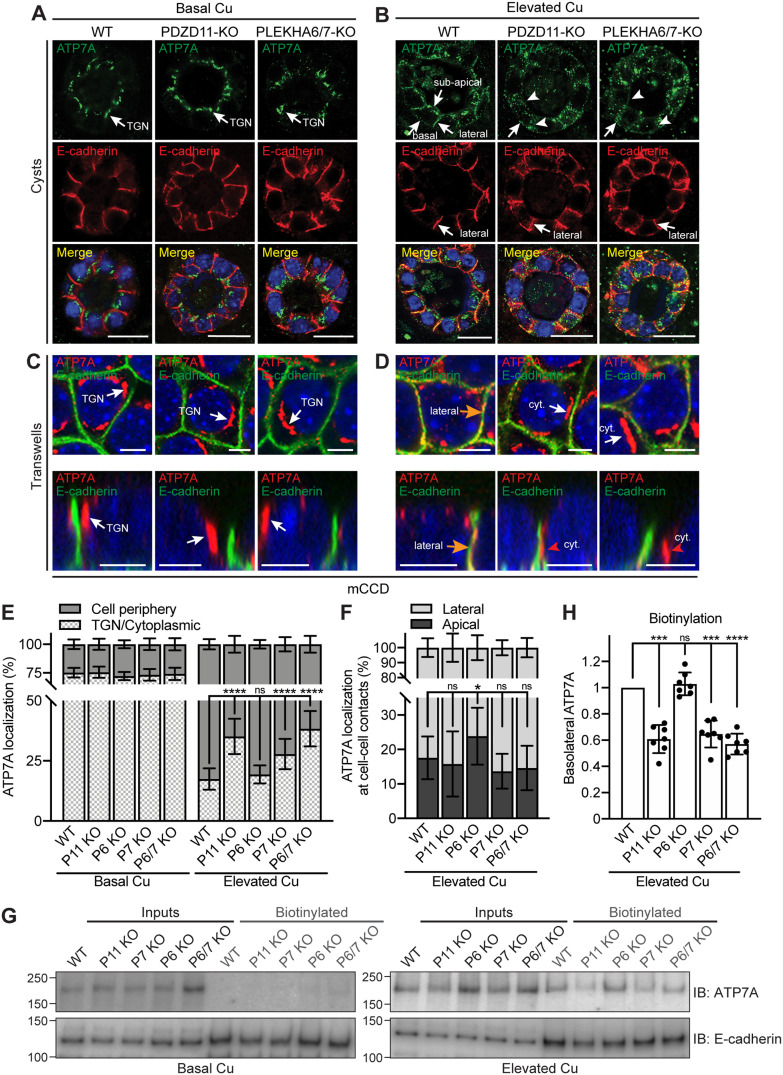
FIGURE 3:
PDZD11, PLEKHA6, and PLEKHA7 are required for the correct targeting of ATP7A to the cell periphery of mCCD cells in elevated copper. (A, B) IF microscopy of the localization of ATP7A (green) and E-cadherin (basolateral labeling marker, red) in mCCD cysts. TGN = trans-Golgi network. Lateral, subapical, and basal ATP7A labeling in elevated copper are indicated by arrows in B. Arrowheads indicate low/undetectable labeling. Bars = 20 µm. (C–F) IF microscopy (C, D) and quantifications (E, F) of ATP7A localization in mCCD cells grown on Transwells (see Supplemental Figure S6 for images at lower magnification). Orange arrows and red arrowheads in D indicate ATP7A labeling colocalized and noncolocalized with E-cadherin, respectively. Bars = 5 µm. Values of ATP7A distribution between cell periphery and TGN/cytoplasm (E) or between lateral and apical cell–cell contacts (F) are shown as mean ± SD. n = 22–30 cells for Basal Cu and n = 92–107 cells for Elevated Cu (E), n = 18–23 cell–cell contacts (F) (one-way ANOVA with post hoc Dunnett’s test, *p < 0.05, ****p < 0.0001, ns, not significant). (G) IB analysis of biotinylated ATP7A in the indicated WT and KO lines under basal and high copper conditions. E-cadherin was used as positive control for basolateral biotinylation. (H) Quantification of basolateral ATP7A in Elevated Cu indicates for each genotype the biotinylated ATP7A signal normalized to the input, relative to WT cells. Dots show replicates (n = 7), and bars represent mean ± SD. One-way ANOVA with post hoc Dunnett’s test (***p < 0.001, ****p < 0.0001, ns, not significant).