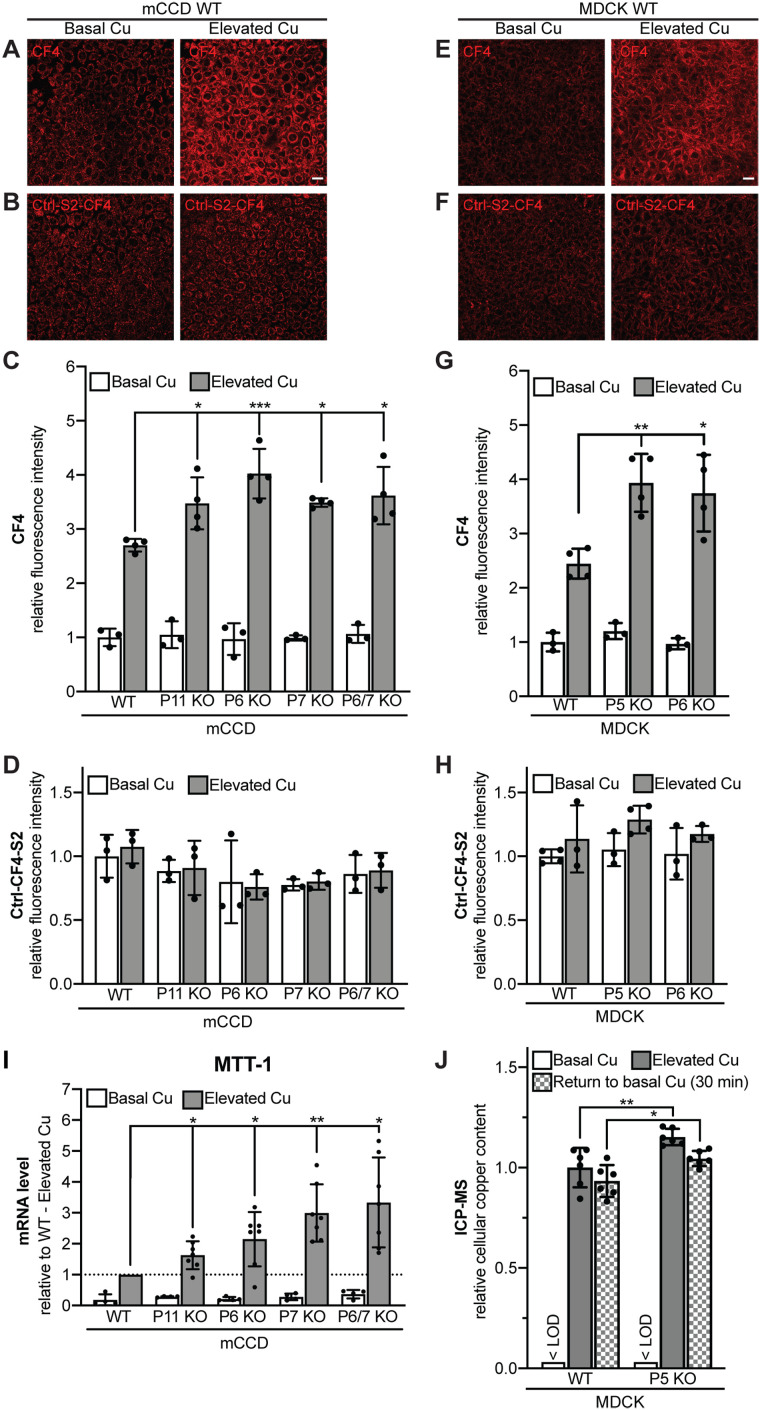
FIGURE 5:
PDZD11 and WW-PLEKHAs are required for the control of labile copper in mCCD and MDCK cells exposed to elevated copper level. Fluorescence images (A, B, E, F) (scale bar = 20 µm), and quantification of fluorescence (C, D, G, H) of either mCCD (A–D) or MDCK (E–H) cells loaded either with CF4 (A, C, E, G) or Ctrl-CF4-S2 (B, D, F, H) under conditions of basal copper (Basal Cu) and after treatment with high concentration of copper (Elevated Cu). In C, D, G, and H, dots show replicates (n), relative to the mean of WT cells in basal copper, and bars represent mean ± SD. One-way ANOVA with post hoc Dunnett’s test (*p < 0.05, **p < 0.01, ***p < 0.001). (I) Quantification by qRT-PCR of the mRNA levels of metallothionein-I (MTT-I) (using GAPDH as internal standard, see Materials and Methods) in WT and KO mCCD cells under conditions of basal copper (Basal Cu) and after treatment with high concentration of copper (Elevated Cu), relative to elevated copper-treated WT cells. Dots show replicates (n = 4 for Basal Cu, n = 7 for Elevated Cu), and bars represent mean ± SD. One-way ANOVA with post hoc Dunnett’s test (*p < 0.05, **p < 0.01). (J) Measurement of cellular copper content by ICP-MS in WT and PLEKHA5 KO MDCK cells under conditions of basal copper (Basal Cu), after treatment with high concentration of copper (Elevated Cu) and after treatment with elevated copper followed by a 30-min treatment in Basal Cu medium (Return to basal Cu) (see Materials and Methods). < LOD = below limit of detection. Dots show replicates (n = 6), relative to the mean of WT cells in elevated copper, and bars represent mean ± SD. One-way ANOVA with post hoc Sidak test (*p < 0.05, **p < 0.01). Abbreviations for genotypes: P11 = PDZD11; P5 = PLEKHA5, P6 = PLEKHA6; P7 = PLEKHA7.