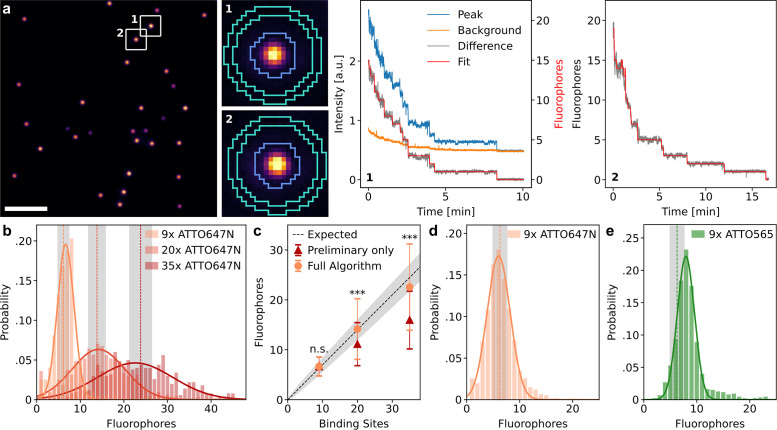
FIGURE 4:
Validation with DNA origami samples. (a) Representative image and traces from the origami experiment with 20 binding sites for ATTO 647N. Scale bar: 10 µm. (b) Fluorophore number distributions for origami with 9, 20, and 35 binding sites. The histograms are modeled with a Gaussian to extract means and SDs (results and sample sizes in Table 1). Vertical dashed lines and areas shaded in gray indicate the expected mean and SD obtained from binomial distributions. (c) Fit results from b compared with the expected mean of the label number distribution, which is a binomial distribution with a labeling efficiency of 70%. Error bars and shaded region show the SD. The quickPBSA result differs significantly from the result without quickPBSA refinement for 20 and 35 binding sites (two sample t test, ***: p < 0.001, n.s.: not significant). (d) Measured label number distribution of origami with nine binding sites for ATTO 647N on a different microscope setup with a larger field of view and sCMOS detector. (e) Measured label number distribution for origami with nine binding sites for ATTO 565.