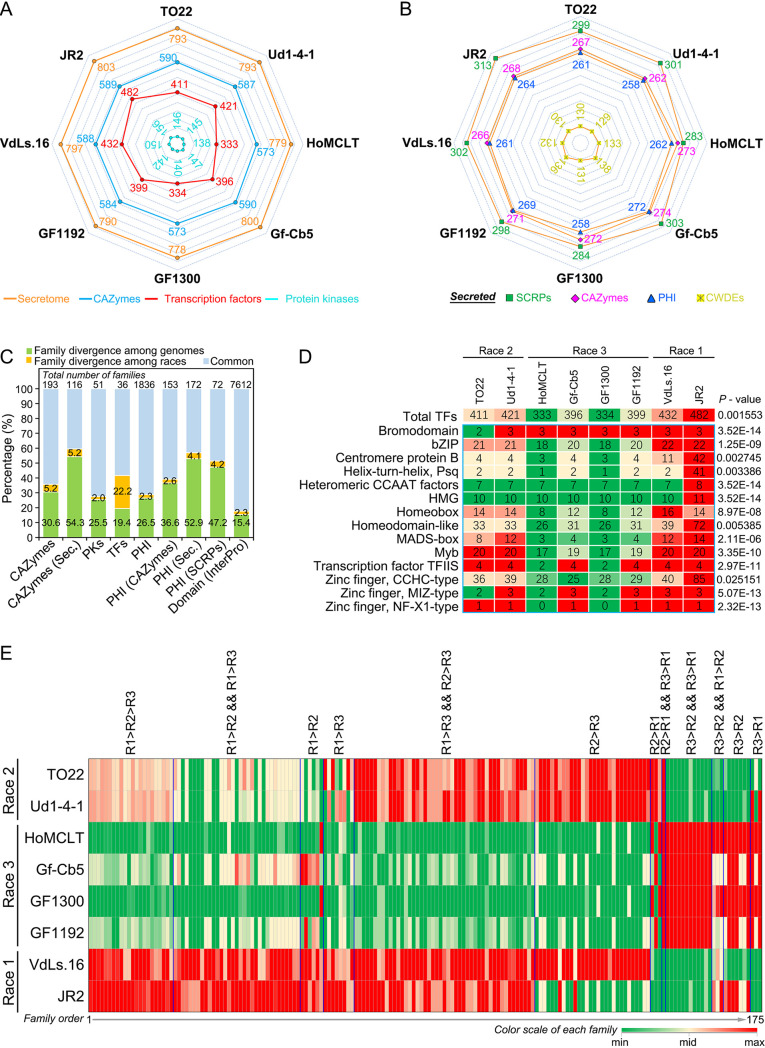
FIG 1.
Functional analysis of the pathogenicity-related gene families among three races in Verticillium dahliae. (A) Comparison of total genes encoding pathogenicity-related proteins among the sequenced genomes, including those encoding secretory proteins, CAZymes, transcription factors, and protein kinases. JR2 and VdLs.16 are the two reference genomes. (B) Number of pathogenicity-related genes encoding secreted proteins among the sequenced genomes. (C) Divergence of the gene families among the sequenced genomes and three races. Gene families in common are represented by equal numbers of genes among the sequenced genomes; family divergence among genomes is represented by the number of genes of a certain race or from any two races that has expanded or contracted relative to the other races; PKs, protein kinases; TFs, transcription factors; PHI, pathogen-host interaction homologs; SCRPs, small cysteine-rich proteins; InterPro, the database used for conserved domain prediction; Sec., encodes protein with secretory characteristics. (D) Transcription factor divergence among three races of Verticillium dahliae. The significant divergence of each subfamily was detected by F test compared to the total number transcription factors among sequenced genomes. (E) Investigation of the functional divergence of predicted proteins by conserved domain (prediction by InterPro database) among the three races. R1, R2, and R3 represent race 1, race 2, and race 3, respectively; “&&” represents that both comparisons are true.