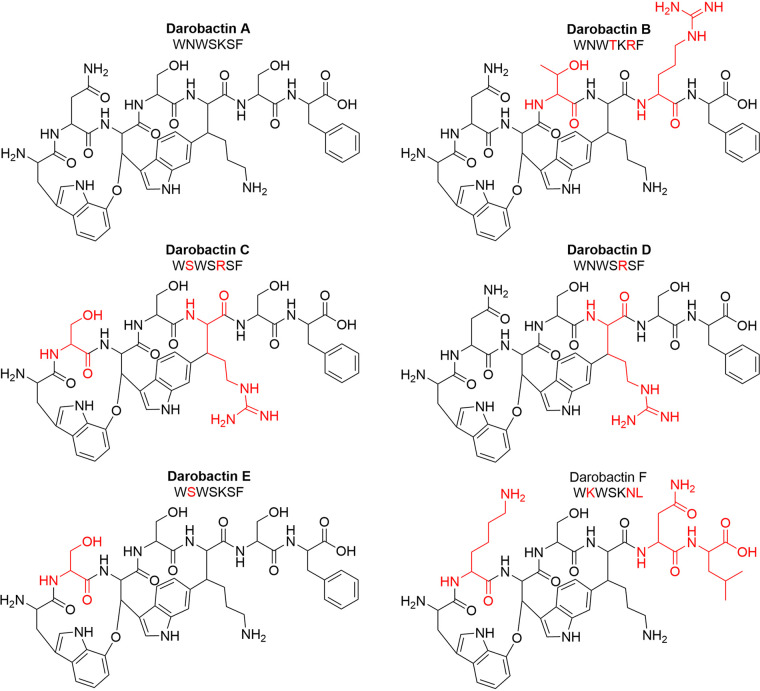
FIG 1.
Structures of in silico-identified darobactin analogs. The amino acid sequences of the bicyclic heptapeptides are given. Predicted producer strains are of the genera Photorhabdus for DAR A, B, and F and Yersinia for DAR C, D, and E. The amino acids that differ from the DAR A sequence are highlighted in red. Exact masses and m/z values for [M + 2H]2+ (in brackets, as mostly the doubly charged ions were observed) for each of the structures are as follows: DAR A = 965.4032 Da (483.7089 m/z), DAR B = 1,048.4879 Da (525.2512 m/z), DAR C = 966.3984 Da (484.2065 m/z), DAR D = 993.4093 Da (497.7119 m/z), DAR E = 938.3923 Da (470.2034 m/z), and DAR F = 972.4818 Da (487.2482 m/z).