Figure 4. Engineering 3′ Cleavage Positions.
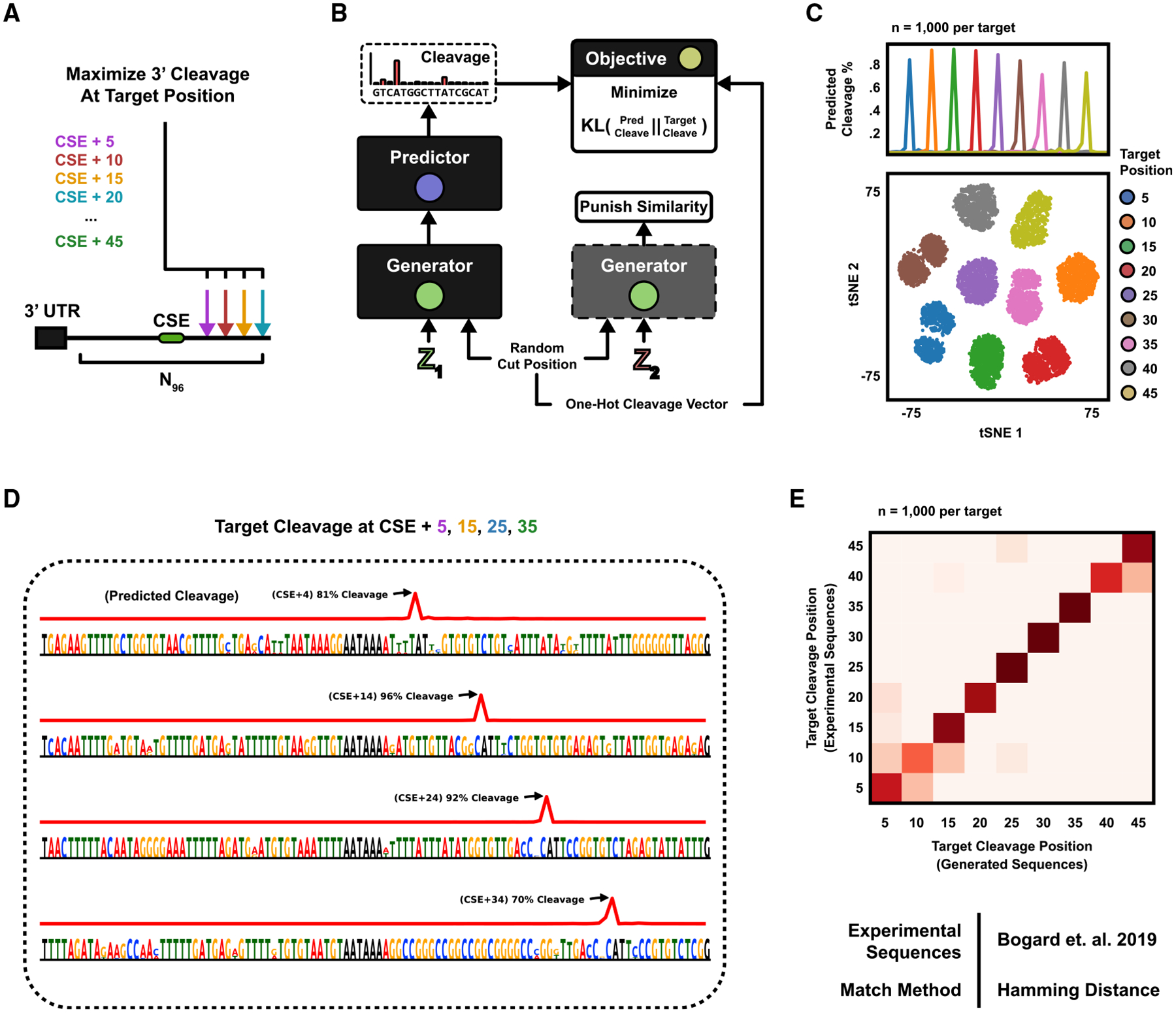
(A) The 3′ mRNA cleavage position is governed by a cis-regulatory code within the PAS. The generative task is formulated as designing PASs, which maximize cleavage at target nucleotide positions downstream of the central hexamer (CSE) AATAAA.
(B) A class-conditional DEN is optimized to generate sequences that are predicted to cleave according to a randomly sampled target cut position. The sampled cut position is also supplied as input to the generator.
(C) The DEN was trained to generate sequences with maximal cleavage at 9 positions, +5 to +45 nt downstream of the CSE, with 50% similarity margin. (Top) Mean predicted cleavage profile (n = 1,000 sequences per target position). Predicted versus target cut position R2 = 0.998. (Bottom) All 9,000 sequences were clustered in tSNE and colored by target position.
(D) Example sequences generated for target positions +5, +15, +25, and +35. 0% duplication rate (n = 100,000 sequences). Hexamer entropy = 9.07 of 12 bits.
(E) The newly generated sequences were compared against gradient ascent-generated sequences for the same target (Bogard et al., 2019) by defining each cluster centroid as the mean one-hot pattern of all DEN-generated sequences and assigning each gradient ascent-pattern to the closest centroid on the basis of L1 distance. Agreement = 0.87.
