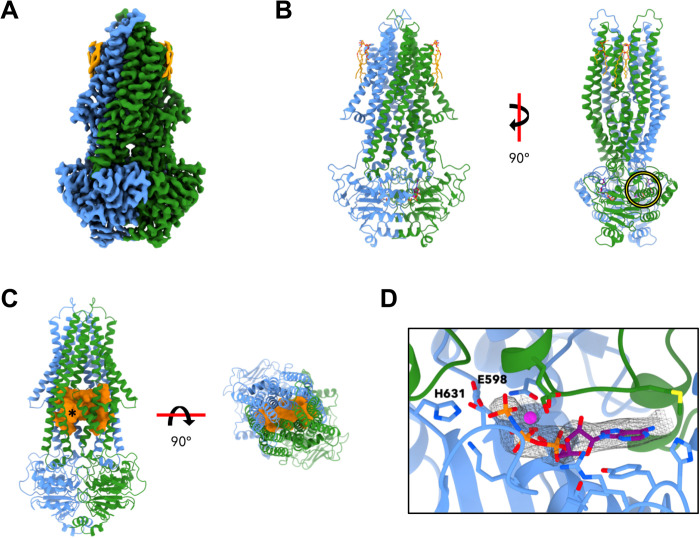
Fig. 2. Cryo-EM structure of Atm1 in MSP1E3D1 nanodiscs with bound AMP-PNP-Mg2+.
(A) The 2.9-Å cryo-EM map reveals an occluded conformation of Atm1 trapped in a prehydrolysis state by addition of AMP-PNP-Mg2+ (color coding as in Fig. 1). (B) The model comprises two bound AMP-PNP molecules (purple) and Mg2+ ions (magenta) at the dimer interface of the NBDs, and two lipids per monomer (orange). A yellow circle identifies the ATP-binding site presented in greater detail in (D). (C) The cavity (orange) is lined by the TM helices and extends across the dimer [left: turned by 40° with respect to (B); right: viewed from the intermembrane space]. At the membrane surface on the matrix side, the asterisk indicates a narrow passage to the matrix. (D) Enlargement of encircled region in (B), with bound Mg2+ and AMP-PNP (gray mesh). Protein color coding as in Fig. 1. The side chains lining the ATP-binding site are shown as sticks. The Walker B E598 mutated in Atm1 E598Q and the switch histidine H631 are pointed out.