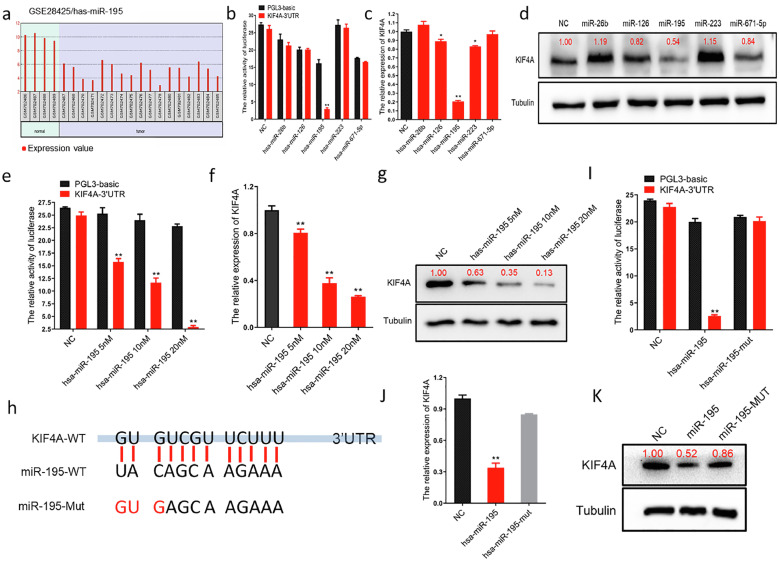
Figure 3.
miR-195 inhibits the expression of kinesin superfamily protein 4A (KIF4A) in osteosarcoma (OS) cells by targeting the KIF4A 3’ untranslated region (UTR).
(a) Analysis of the Gene Expression Omnibus (GEO) database-derived dataset GSE28425. Bar charts showed that the miR-195 expression level was significantly lower in OS tissue (n=4) than in normal tissue (n=17) (p<0.01). (b) The targeted binding effect between the KIF4A 3′ untranslated region (UTR) and miRNAs determined using a luciferase assay in HEK293 cells (Black columns represent the luciferase activity co-transfected with PGL-3 basic; Red columns represent the luciferase activity co-transfected with KIF4A 3′UTR, **p<0.01). (c) The inhibitory effect of miRNAs on KIF4A detected using qPCR in MG63 cells. miR-195 mimic inhibited the expression level of KIF4A in MG63 cells (**p<0.01). (d) The inhibitory effect of miRNAs on KIF4A detected using western blotting in MG63 cells. (e) Luciferase dose-dependent assay showing significant binding of miR-195 to the 3’UTR region of KIF4A in HEK293 cells (Black columns represent the luciferase activity co-transfected with PGL-3 basic; Red columns represent the luciferase activity co-transfected with KIF4A 3′UTR, **p<0.01). (f) qPCR dose-dependent assay showing that miR-195 inhibited the expression of KIF4A in MG63 cells (**p<0.01). (g) western blotting dose-dependent assay showing that miR-195 inhibited the expression of KIF4A in MG63 cells. (h) Potential binding sites between miR-195 and KIF4A were predicted with miRNA target prediction software including BiBiServ, MIRDB, and DIANA.. (i) The effect of miR-195 point mutations on KIF4A binding determined using a luciferase assay in HEK293 cells (**p<0.01). (j) The effect of miR-195 point mutations on KIF4A binding detected using qPCR in MG63 cells. (k) The effect of miR-195 point mutations on KIF4A binding detected using western blotting in MG63 cells.