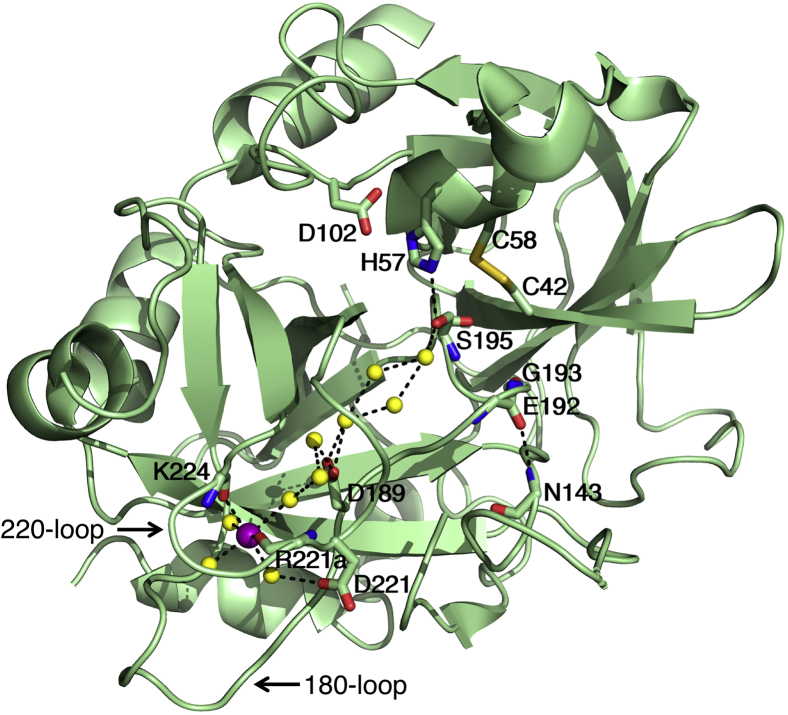
Figure 1.
Ribbon representation of thrombin in the E form bound to Na+(PDB ID:1SG8) (17). The enzyme folds as typically observed for proteases of the trypsin family and is depicted in the Bode orientation, with the active site wide open at the center and the 180 and 220 loops in the south-west corner (60). The residues of the catalytic triad D102/H57/S195 are shown with relevant H-bonds. Na+ (purple) is bound within these loops and coordinated by four buried water molecules and two backbone O atoms from R221a and K224 (side chains not shown for clarity). D189 at the bottom of the specificity pocket participates indirectly in the coordination shell by supporting one of the waters through H-bonding, a function also played by the side chain of D221. An important backbone H-bond between N143 and E192 (side chains not shown for clarity) stabilizes the orientation of the backbone N atoms of S195 and G193 that define the oxyanion hole responsible for coordination of substrate in the transition state (1, 2, 3). A H-bonded network of water molecules (yellow) connects the Na+ site to the Oγ atom of S195 across a distance of >15 Å. The position of the C48/C52 disulfide bond next to the catalytic S195 is noted.