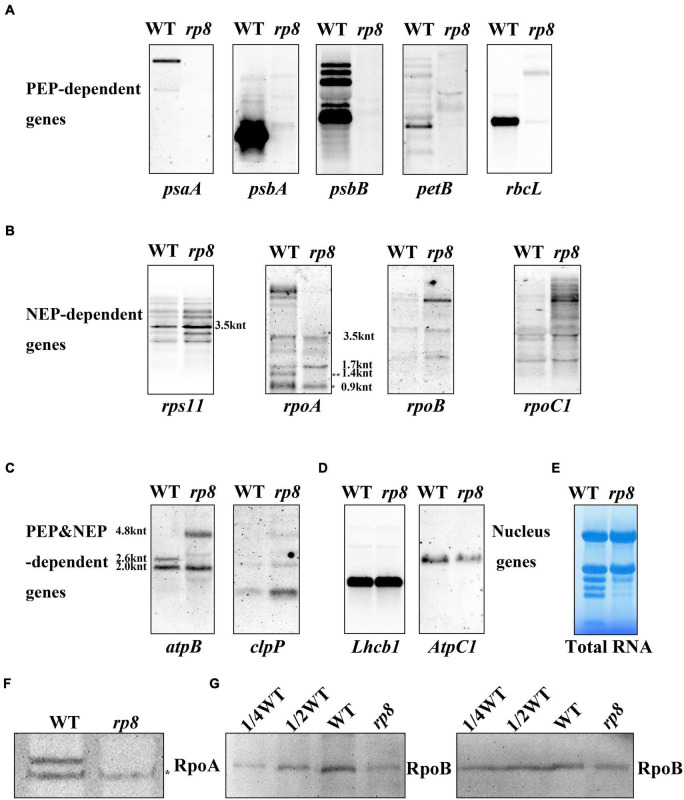
FIGURE 6.
Northern blot analysis of chloroplast-associated genes in the wild type (WT) and the rp8 mutant. (A) Steady-state levels of PEP-dependent transcripts, psaA, psbA, psbB, petB, and rbcL. (B) Steady-state levels of NEP-dependent transcripts (rps11, rpoA, rpoB, and rpoC1). * Indicates the mature transcripts of rpoA (990 nt), ** indicates the 1.4 knt transcript as described in Zhang et al. (2015). The sizes of the transcripts were indicated according to Wu and Zhang (2010). (C) Steady-state levels of chloroplast transcripts that are both PEP- and NEP-dependent (atpB and clpP). As for the atpB transcripts, 4.8, 2.6, and 2.0 knt transcripts are indicated. (D) Steady-state levels of two nucleus genes (Lhcb1 and AtpC1). (E) Methylene blue staining of total RNA for loading control. About 10 μg total RNA was transferred to a nylon membrane after electrophoresis, then probed with DIG-labeled probes. (F) Immunoblot analysis of RpoA with the antibody described in Zhang et al. (2018) (*represents the specific band for the RpoA protein). (G) Immunoblot analysis of RpoB protein levels in the wild type (WT) and the rp8 mutant. Total proteins from the WT samples were loaded with three different concentrations (7.5, 15, and 30 μg), and total proteins from the mutant samples were loaded with 30 μg, respectively. Arabidopsis mutant ptac10 with a decreased RpoB protein level was used as a control (Chang et al., 2017; Yu et al., 2018).