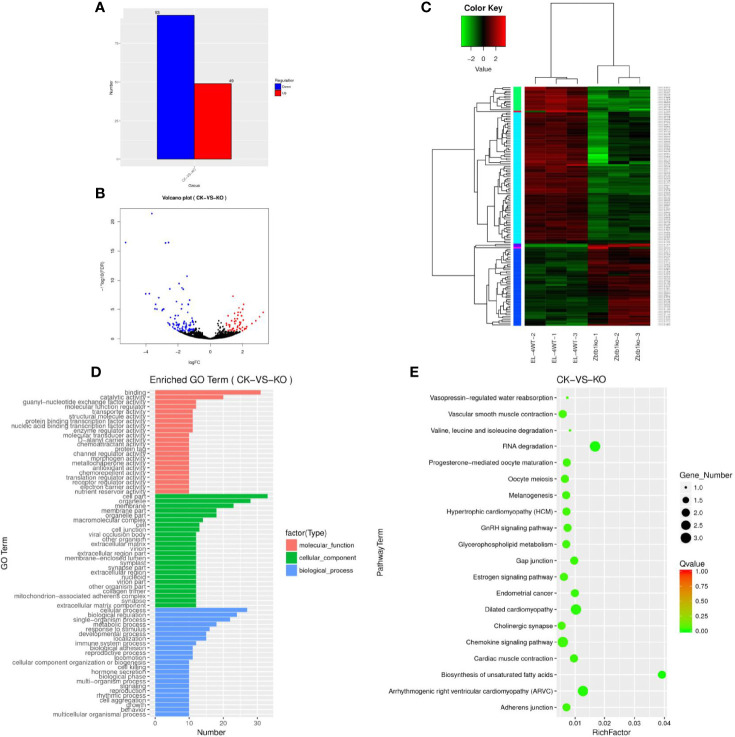
Figure 1.
(A) Sample difference comparing the upregulation and downregulation of lncRNA gene expression. (B) Volcano map of differential lncRNA genes. Red dots indicate significantly upregulated genes, and blue dots indicate downregulated expression. The abscissa represents multiple changes in gene expression in different samples; the ordinate represents statistically significant changes in gene expression. (C) Differential lncRNA gene cluster map clustered by log10 (FPKM+1) values; red indicates highly expressed genes, and green represents genes with low expression. As the color shifts from red to blue, a higher gene expression is indicated. (D) The differential lncRNA target gene GO enrichment histogram and the ordinate enrichment GO term. The abscissa is the number of differential genes in the term. Different colors are used to distinguish biological processes, cellular components and molecular functions. (E) KEGG enrichment and scatter map of differential lncRNA target genes. The vertical axis represents the pathway name, the horizontal axis indicates the size of the rich factor, the dot indicates the number of differentially expressed genes in this pathway, and the color of the dot corresponds to different Q value ranges.