Fig. 1.
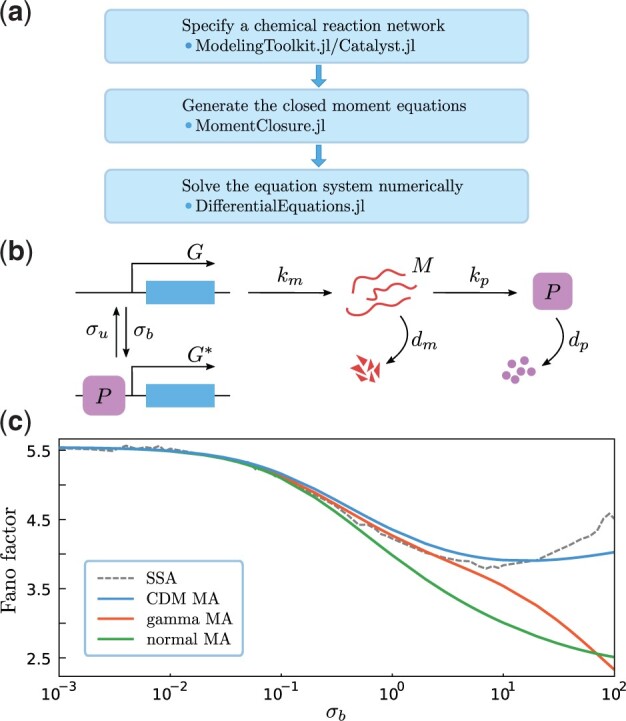
(a) The general workflow of moment-based modelling in Julia using MomentClosure.jl and related packages. (b) Model of a negative auto-regulative genetic feedback loop. If a gene is active (G), an mRNA molecule (M) is produced with rate km which can be subsequently translated into proteins (P) with rate kp (both degrade with rates dm and dp, respectively). The negative feedback is introduced via protein binding to the gene with rate σb, switching the promoter OFF () and preventing the transcription (in contrast, switching ON occurs with rate σu). (c) Fano factor of the steady-state protein number as a function of σb where we have truncated the moment hierarchy at the second order using normal, gamma and CDM MAs, and compared the results to the true values predicted by the stochastic simulation algorithm (Gillespie, 1977) (averaged over 105 realizations). The initial condition is zero protein and mRNA molecules (set to 0.001 for MAs to ensure numerical stability) in state G, and the parameters are fixed as and dp = 1
