Abstract
The emergence and spread of antimicrobial resistance (AMR) among bacterial pathogens have created a global threat to human health and the environment. Targeting the quorum sensing (QS) linked virulent traits of bacteria is considered to be a novel approach for addressing the problem of AMR. In this study, green synthesized silver nanoparticles (AgNPs-MK) were evaluated for the inhibition of the formation of biofilms and quorum sensing controlled virulence factors against three Gram negative bacteria. Remarkable inhibition (>80%) of QS-mediated violacein production was recorded in C. violaceum 12472. Up to 90% inhibition of the QS-mediated virulent traits of S. marcescens MTCC 97 was observed. The virulence factors of P. aeruginosa PAO1 also decreased in a dose dependent manner in the presence of AgNPs-MK. Moreover, the development of biofilms of C. violaceum 12472, S. marcescens MTCC 97, and P. aeruginosa PAO1 was reduced by 87.39, 81.54, and 71.34%, respectively. Biofilms on glass surfaces were remarkably reduced, with less aggregation of bacterial cells and the reduced formation of extra polymeric substances. The findings clearly show the efficacy of AgNPs-MK against the development of biofilms and the QS mediated virulent traits of Gram negative bacterial pathogens. AgNPs-MK may be further exploited for the development of alternative antimicrobial agents after careful scrutiny in animal models for the management of bacterial infections, especially for topical applications.
Green synthesized silver nanoparticles inhibit the quorum sensing and biofilm development of Gram-negative pathogenic bacteria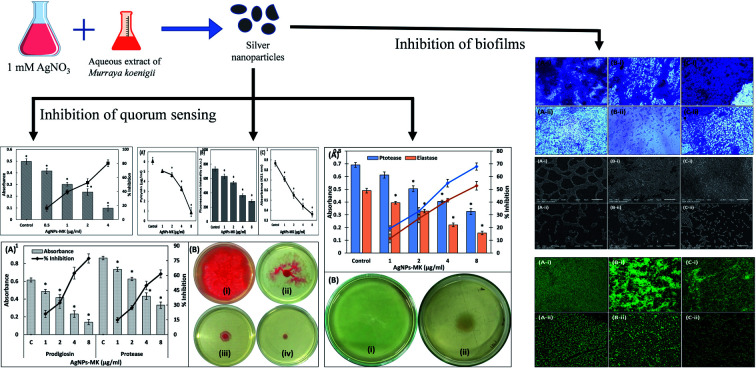
1. Introduction
In the last few decades, there has been a tremendous increase in the incidence and emergence of multi-drug resistance (MDR) among bacterial pathogens, both in developing and developed countries.1 Today, the diseases caused by infectious microbes have become the major cause of global mortality and morbidity after cancer and cardiovascular diseases.2 These MDR pathogens or “superbugs” are now considered to be an epidemiological concern and antimicrobial resistance (AMR) worsens the treatment of infectious diseases by reducing the therapeutic efficacy of antibiotics.3,4 The situation has become so alarming that if no action is taken, AMR will become a major cause of mortality, even surpassing cancer in the next few decades.5,6 AMR not only poses a burden on public health systems, but is also problematic for livestock and the environment. The development of AMR has also complicated the management of other chronic diseases. The first six or seven decades of the 20th century is considered as the golden era for the discovery of antibiotics and this period resulted in the discovery of 70% of all antibiotics reported to date. A timeline of antibiotic discovery shows a large gap or void for almost four decades, during which only a few antibiotics were discovered that exhibit a novel mechanism of action.7 The antibiotic selection pressure is one of the key drivers in the development of AMR among microbial pathogens.8,9 The selection pressure is developed by the sub-judicious and unprescribed use of antibiotics in human health care, livestock and the environment. Now, even the latest generation of antibiotics can no longer be trusted for long term applications because of the risk of development of resistance. Therefore, there are two major problems that need to be addressed in antibacterial drug discovery; one is the development of new therapeutic antimicrobials with a novel mode of action, and the other is to prevent the development of AMR against the discovered antimicrobials. This has created a need for the development of alternative anti-infective strategies to combat AMR.
Targeting quorum sensing (QS) regulated virulence factors and biofilm development are considered as promising anti-infective drug targets to combat the AMR problem.10 QS is a microbial cell to cell communication system in which microbes express their certain genes only when their population reaches a certain threshold limit. Bacteria synthesize and secrete certain signal molecules called autoinducers (AIs). The concentration of AIs increases with the increase in the bacterial population. When the population density of bacteria reaches a certain limit, a certain set of genes are expressed by transcriptional regulation.11 The AIs in Gram negative bacteria are N-homoserine lactones (AHLs), and autoinducer (AI) peptides in Gram positive bacteria. Many virulent traits of bacteria, as well as the production of antibiotic degrading enzymes, are controlled via QS. The previous assumption about the growth of bacteria was that they live in a planktonic state, but later it was found that most bacteria live in complex structures called biofilms. Biofilms are comprised of microbial communities and extracellular polymeric substances (EPS) that act as a protective barrier.12 In the biofilm mode of growth, the expression of some phenotypes of bacteria are different from those in planktonic growth. The importance of biofilms can be understood by considering that the National Institute of Health (NIH) estimates that nearly 80% of microbial infections are encouraged and established by biofilms.13,14 The vast majority of infections are caused by the development of biofilms, either by pathogenic or opportunistic pathogens.15,16
A new approach in the discovery of antimicrobials is to target the bacterial QS and biofilms using green synthesized nanoparticles. Previous reports have indicated that silver nanoparticles may prove to be useful as alternative antimicrobial agents.17 Nanotechnology has gained vast attention in scientific research owing to the possibly of its application in medicine, bioremediation, diagnostics, agriculture and so on.18 Many materials produce better biological effects in the nano form compared to their bulk state, mainly because they exhibit different physical and chemical properties at this scale. It is anticipated that nanotechnology has possible applications in many disciplines of health care, such as for use as novel drugs, in drug delivery, diagnostics, and improved biomaterials (medical devices). There are certain risks associated with the use of nanoparticles in medicine that include toxicity to the host's system. There are numerous approaches for the synthesis of metal nanoparticles, such as biological, chemical, and physical methods. The major disadvantages of the chemical methods are the formation of hazardous and non-biodegradable byproducts that are toxic to the environment. The ‘green synthesis’ approach is gaining significant attention as it minimizes waste production and uses safer (non-toxic) solvents/materials, making this approach more environmentally friendly.19
Previously, many studies have demonstrated the efficacy of green synthesized nanoparticles against drug resistant bacteria.17 In this study, the aqueous extract from M. koenigii was used to synthesize silver nanoparticles (AgNPs-MK). The effect of AgNPs-MK was tested for the broad-spectrum inhibition of biofilm formation and QS mediated virulence factors of three Gram negative bacteria, viz. C. violaceum 12472, P. aeruginosa PAO1, and S. marcescens MTCC 97.
2. Materials and methods
2.1. Materials and reagents
Azocasein and elastin congo red (ECR) were purchased from Sigma Aldrich, USA. Trichloroacetic acid (TCA), orcinol and bacteriological culture media were procured from HiMedia Laboratories, Mumbai, India. 2,3,5-Triphenyltetrazolium chloride (TTC) was purchased from SRL Pvt. Ltd.
2.2. Synthesis of AgNPs-MK
The silver nanoparticles (AgNPs-MK) were synthesized using an aqueous extract of Murraya koenigii. Detailed information on the AgNPs-MK synthesis and the characterization data has been published previously.20 Briefly, an aqueous extract of M. koenigii leaves was prepared by dissolving 5 g plant material in 100 ml double distilled water. For the synthesis of the silver nanoparticles, 0.5 ml of the extract was mixed with 20 ml AgNO3 solution (1 mM) and placed on a magnetic stirrer for 4 h. The colour of the reaction mixture changed to dark brown indicating the formation of silver nanoparticles. The nanoparticles (MK-AgNPs) were harvested by centrifugation and washed thrice with distilled water. The particles were then dried in an oven at 60 °C and stored for further use. The nanoparticles were characterized using UV-vis spectroscopy, X-ray diffraction (XRD), Fourier-transform infrared spectroscopy (FTIR), transmission electron microscopy (TEM), scanning electron microscopy, and energy dispersive X-ray (EDX) analysis. The addition of the extract to silver nitrate solution resulted in a colour change of the reaction mixture. AgNPs-MK exhibited a surface plasmon resonance (SPR) band at 410 nm, this indicated the formation of silver nanoparticles. TEM analysis revealed that the size of the AgNPs-MK ranged from 5 to 20 nm. Moreover, most of the nanoparticles were spheroidal in shape and a few nanoparticles exhibited an anisotropic morphology. The average size of the AgNPs-MK calculated from the XRD data was 13.54 nm.
2.3. Determination of minimum inhibitory concentrations
The minimum inhibitory concentration (MIC) of the AgNPs-MK against tested bacterial strains was determined using the microbroth dilution method using TTC as a growth indicator dye.21 The AgNPs-MK (10 μl) were added to 190 μl Luria Bertani broth in a 96-well microtiter plate and were diluted two-fold to give treatments of varying concentrations. The diluted culture (100 μl, 1 : 50) of bacteria from the log phase was added as an inoculum in each well. The microtitre plate was incubated overnight at the respective optimum growth temperatures of each bacterium. TTC (20 μl, 2 mg ml−1) was added to each well and the plate was incubated at 37 °C for 30 min in the dark. The wells were examined for colour change. The development of a pink/red colour indicated the presence of actively growing cells, while no colour change showed a lack of bacterial growth. Wells showing no change in colour were spotted on Luria-Bertani (LB) agar plates to further confirm the growth inhibition.
2.4. Quantitative determination of the violacein pigment in C. violaceum 12472
The quantitative assessment of violacein production was performed following the standard procedure.22 In short, C. violaceum 12472 was grown in the absence and presence of varying sub-MICs of AgNPs-MK for 18 h at 30 °C. The grown culture (1 ml) was centrifuged (10 000 rpm) for 5 min to separate out the insoluble pigment (violacein) and microbial cells. The pellet obtained was resuspended in DMSO in a volume of 1 ml and vortexed vigorously (5 min) to dissolve the pigment. The suspension was again centrifuged to spin down the debris from the bacterial cells. The supernatant was collected and its absorbance was recorded at 585 nm using a UV-2600 spectrophotometer, Shimadzu, Japan.
2.5. Assays for the inhibition of the virulence factors of S. marcescens MTCC 97
Various QS regulated virulence factors of S. marcescens MTCC 97 were determined in the presence and absence of AgNPs-MK at sub-minimum inhibitory concentrations (MICs). The assays used for determination of individual virulence factors are briefly described below.
2.5.1. Assessment of prodigiosin production
The assessment of the prodigiosin pigment was carried out in Luria-Bertani medium following the standard method.23 Briefly, S. marcescens MTCC 97 was cultured in the absence and presence of sub-MICs of AgNPs-MK for 18 h at 30 °C. 2 ml of the grown culture was centrifuged at 10 000 rpm for 5 min to pellet down the bacterial cells. The pellet was dissolved in 1 ml of acidified ethanol (96 ml ethanol + 4 ml 1 M HCl) by vigorous vortexing for 5 min. The sample was again centrifuged to remove the debris and the absorbance of the supernatant was recorded at 534 nm using a UV-2600 spectrophotometer.
2.5.2. Exoprotease activity
The exoprotease activity of S. marcescens MTCC 97 was determined using a azocasein degradation assay.24S. marcescens MTCC 97 was grown in the absence and presence of sub-MICs of AgNPs-MK for 18 h at 30 °C. The culture supernatant was obtained by centrifugation and the supernatant (0.1 ml) was mixed with 1 ml of 0.3% (w/v) azocasein (0.5 mM CaCl2 in 50 mM M Tris- HCl, pH 7.5). The reaction mixture was incubated for 15 min at 37 °C under shaking conditions. The reaction was terminated by adding 0.5 ml of 10% ice cold trichloroacetic acid, followed by centrifugation to remove the insoluble azocasein. The absorbance of the supernatant was recorded at 400 nm using a UV-2600 spectrophotometer.
2.5.3. Determination of the swimming motility of S. marcescens MTCC 97
Briefly, 5 μl of the culture from the log phase of S. marcescens MTCC 97 was spotted on LB plates (0.5% agar) containing varying sub-MICs of AgNPs-MK. No amendments were performed on the control plates. The plates were incubated for 18 h at 30 °C. The swimming zone was measured using a transparent ruler in millimetres (mm).
2.6. Determination of the virulence factors of P. aeruginosa PAO1
Various QS regulated virulence factors of P. aeruginosa PAO1 were examined in the absence and presence of AgNPs-MK. The methods used for determination of individual virulence factors are briefly described below.
2.6.1. Estimation of pyocyanin
The assessment of the pyocyanin pigment was carried out in Pseudomonas broth (PB) as this medium enhances the production of the pyocyanin pigment.25 Briefly, P. aeruginosa PAO1 was cultured in the absence and presence of sub-MICs of AgNPs-MK for 18 h at 37 °C. 5 ml of the grown culture was centrifuged at 10 000 rpm for 5 min to pellet down the bacterial cells. Pyocyanin present in the cell free supernatant was extracted in chloroform (3 ml) by vortexing and the aqueous phase was discarded. The organic (chloroform) phase was re-extracted in 1.2 ml of 0.2 N HCl. The absorbance of the aqueous phase was recorded at 520 nm using a UV-2600 spectrophotometer against 0.2 N HCl as blank.
2.6.2. Assessment of pyoverdin
The pyoverdin levels were assessed spectrophotometrically following the standard procedure.26P. aeruginosa PAO1 was cultured overnight at 37 °C in the absence and presence of sub-MICs of AgNPs-MK. A cell free supernatant was obtained by centrifugation. 0.1 ml supernatant was mixed with 0.9 ml of 50 mM Tris–HCl (pH 7.4). The fluorescence emission signal of the sample was measured (460 nm) by excitation at 400 nm using a RF-5301PC spectrofluorometer (Shimadzu, Japan).
2.6.3. Assessment of rhamnolipid
The rhamnolipid level was determined in the control and treated group using the orcinol method as described previously.27 Briefly, P. aeruginosa PAO1 was grown for 18 h at 37 °C in the absence and presence of sub-MICs of AgNPs-MK. The cell free supernatant was obtained by centrifugation. The supernatant (300 μl) was mixed with 600 μl diethyl ether and vortexed for 1 min. The mixture was left to enable separation under static conditions and the organic phase was removed. The organic phase was completely dried by evaporating at room temperature and then reconstituted in 0.1 ml deionized water. Orcinol solution (0.9 ml, 0.19% w/v orcinol in 53% H2SO4) was added to each sample and the reaction mixture was heated for 30 min at 80 °C. The samples were cooled at room temperature and the absorbance was recorded at 421 nm using a UV-2600 spectrophotometer.
2.6.4. Exoprotease activity
The exoprotease activity in the supernatant of P. aeruginosa PAO1 was determined using an azocasein degradation assay.27P. aeruginosa PAO1 was grown for 18 h at 37 °C in the absence and presence of sub-MICs of AgNPs-MK followed by isolation of the cell free supernatant by centrifugation. The culture supernatant (0.1 ml) was mixed with 1.0 ml of 0.3% (w/v) azocasein (prepared in 0.05 M Tris–HCl containing 0.5 mM CaCl2, pH 7.5). The reaction mixture was incubated for 4 h at 37 °C under shaking conditions and the reaction was terminated by adding 0.5 ml ice cold trichloroacetic acid (10% w/v). The reaction mixture was centrifuged (13 000 rpm for 5 min) to remove the insoluble azocasein and the absorbance of the supernatant was recorded at 400 nm using a UV-2600 spectrophotometer.
2.6.5. Assessment of the LasB elastinolytic activity
The elastinolytic activity in P. aeruginosa PAO1 was evaluated using the standard Elastin Congo Red (ECR) assay.28P. aeruginosa PAO1 was grown in the absence and presence of sub-MICs of AgNPs-MK for 18 h at 37 °C. Cell free supernatant was obtained by centrifugation. 0.1 ml supernatant was mixed with 0.9 ml ECR buffer (5 mg ml−1 ECR and 1 mM CaCl2 in 100 mM Tris, pH 7.5) and incubated at 37 °C for 3 h under shaking conditions. The reaction was stopped by the addition of 1 ml of sodium phosphate buffer (100 mM, pH 6.0) and placing the samples at 4 °C for 30 min. The insoluble ECR was separated by centrifugation (13 000 rpm for 5 min) and the absorbance was recorded at 495 nm using a UV-2600 spectrophotometer.
2.6.6. The swimming motility of P. aeruginosa PAO1
Briefly, a 5 μl culture from the log phase of P. aeruginosa PAO1 was spotted on LB plates (0.3% agar) containing varying sub-MICs of AgNPs-MK. No amendments were made to the control plates. The plates were incubated for 18 h at 37 °C under static conditions. The swimming zone was measured using a transparent ruler in millimetres (mm).
2.7. Assays to measure biofilm inhibition
2.7.1. Assessment of biofilm in a 96-well microtitre plate
Quantitative evaluation of biofilm inhibition by AgNPs-MK was determined using a crystal violet assay in a 96-well microtitre plate following the standard method.29 Cultures of test bacteria grown overnight were inoculated in the wells of a microtitre plate containing 150 μl LB medium. Varying sub-MICs of AgNPs-MK were added and control wells were not subjected to any treatment. The plates were incubated for 24 h at the respective bacterial growth temperatures under static conditions. The excess broth and planktonic cells were removed by washing three times with sterile phosphate buffer and the wells were air dried for 20 min at room temperature. The biofilms were then stained with 200 μl of crystal violet solution (0.1% w/v) for 15 min. The wells were again washed gently to remove the unbound dye. The biofilms bound to crystal violet were dissolved in 200 μl 90% ethanol. The absorbance of the sample was recorded at 620 nm using a microplate reader (Thermo Scientific Multiskan EX, UK).
2.7.2. The microscopic evaluation of biofilm inhibition
2.7.2.1. Light microscopy
Briefly, 60 μl of cultures of test bacteria grown overnight were inoculated into 24-well culture plates containing 3 ml of culture media. Sterile glass coverslips (1 × 1 cm) and the respective highest sub-MICs of the AgNPs-MK were added to the wells. The control group was not subjected to any treatment. After 24 h of incubation, the loosely bound cells were removed from the coverslips by gentle washing with phosphate buffer and air dried for 20 min at room temperature. The coverslips containing the biofilms were stained with 0.1% w/v crystal violet solution for 15 min. The excess dye was washed away and the slides were finally air dried for 30 min. The biofilms were visualized under a light microscope (Olympus BX60, Model BX60F5, Olympus Optical Co. Ltd Japan) equipped with a colour VGA camera (Sony, Model no. SSC-DC-58AP, Japan). The images were captured at 40× magnification.
2.7.2.2. Confocal laser scanning microscopy
The biofilms were formed on glass coverslips in the absence and presence of AgNPs-MK as mentioned in the light microscopy section. After 24 h of incubation, the loosely bound cells were removed from the coverslips by gentle washing with sterile phosphate buffer and air dried for 20 min at room temperature. The coverslips were then stained with 0.1% acridine orange for 20 min and air dried for 60 min at room temperature in the dark. The biofilms were visualized using Zeiss LSM780, at University Sophisticated Instrumentation Facility (USIF), AMU, Aligarh, and images were captured at 63× magnification.
2.7.2.3. Scanning electron microscopy
The biofilms were formed on glass coverslips in the absence and presence of AgNPs-MK as mentioned in the light microscopy section. The coverslips were washed to remove loosely adhered cells and air dried. The biofilms were fixed with glutaraldehyde (2.5% in 50 mM phosphate buffer) at 4 °C for 24 h. The cells in biofilms were dehydrated using an increasing gradient (20 to 100%) of ethanol. The glass coverslips were air dried and coated with gold before visualization. The biofilms were visualized and the images were captured using a JEOL-JSM 6510 LV at USIF, AMU, Aligarh.
2.8. Statistical analysis
All experiments were performed as three independent replicates and the data presented is the mean with standard deviation. The statistical significance of the data was determined by calculating the P-value using the t-test with respect to control. A P-value of less than 0.05 with respect to the control was considered as significant and is denoted with *.
3. Results and discussion
3.1. MICs of AgNPs-MK
The MICs of the AgNPs-MK against C. violaceum 12472, P. aeruginosa PAO1, and S. marcescens MTCC 97 were found to be 8, 16, and 16 μg ml−1, respectively. The effect of AgNPs-MK on biofilm formation and QS regulated virulence factors was tested below the inhibitory concentrations (sub-MIC).
3.2. Effect of AgNPs-MK on the QS-mediated virulence factors of C. violaceum 12472
3.2.1. Inhibition of violacein pigment production
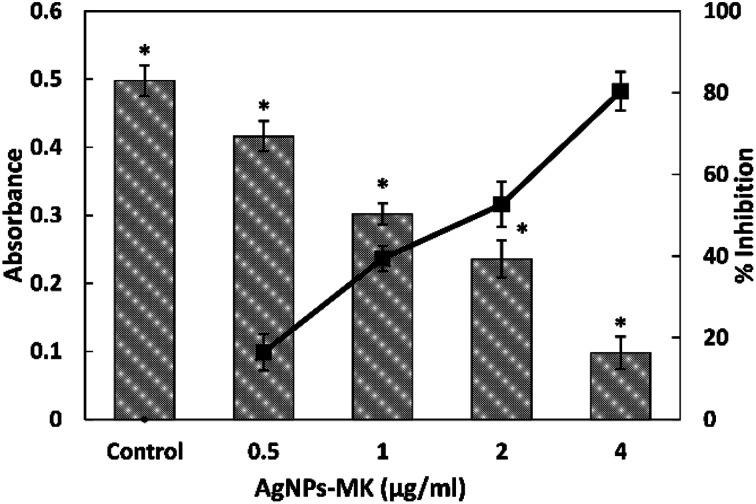
The preliminary anti-QS activity of the AgNPs-MK was determined by assessing its effect on the production of violacein pigment in C. violaceum 12472. This is a test strain in which the pigment production is controlled by QS.30 Any decrease in the pigment production is a reflection of the anti-quorum sensing (anti-QS) activity. The absorbance of the extracted pigment in the control group was found to be 0.497 ± 0.022, which decreases in a dose dependent manner upon treatment with AgNPs-MK as shown in Fig. 1. Treatment with 0.5, 1, 2, and 4 μg ml−1 AgNPs-MK reduced the violacein biosynthesis by 16.40, 39.38, 52.64, and 80.37% in C. violaceum 12472, respectively. The result clearly demonstrates that the green synthesized AgNPs-MK exhibited anti-QS activity. This observation is in agreement with our previous finding, in which silver nanoparticles synthesized using Carum copticum extract reduced violacein pigment production in C. violaceum 12472 by 83.2% at 5 μg ml−1.31
Fig. 1. The quantitative analysis of violacein inhibition in C. violaceum 12472 using AgNPs-MK. Data are represented as average values and the error bars represent the SD. The percentage inhibition is shown on the secondary y-axis. * indicates p ≤ 0.05 with respect to the control.
3.2.2. Inhibition of swimming motility
The anti-QS activity of AgNPs-MK was further tested against another virulence factor of C. violaceum 12472, that is, the swimming motility. The average swim diameter of control C. violaceum 12472 was 27.66 mm. Treatment with AgNPs-MK did not result in any significant inhibition of the swimming motility. There was a 7.22% inhibition at the highest sub-MIC (4 μg ml−1), but it was also statistically insignificant (P-value = 0.43).
3.3. Inhibition of the virulence factors of S. marcescens MTCC 97 by AgNPs-MK
3.3.1. Inhibition of prodigiosin production
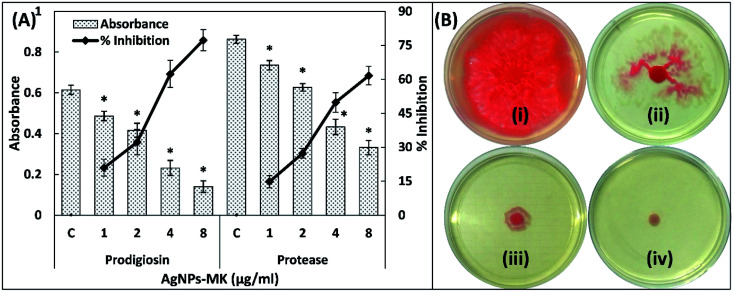
To study the broad-spectrum QS inhibition of the synthesized AgNPs-MK, anti-QS activity was also tested against S. marcescens MTCC 97. Prodigiosin is a pink-red pigment produced by S. marcescens, the synthesis of this pigment is controlled by QS [50]. A minimum four AIs, that is AHLs are synthesized by S. marcescens and control the expression of many virulence factors including the formation of biofilms, prodigiosin production, motility, and carbapenem resistance.32 The presence of varying sub-MICs of AgNPs-MK exhibited the inhibition of prodigiosin production in S. marcescens MTCC 97 as shown Fig. 2A. A 20.92, 32.24, 62.33, and 77.23% inhibition of prodigiosin biogenesis was observed in the presence of 1, 2, 4, and 8 μg ml−1 of AgNPs-MK, respectively. A previously published study reported that some virulence factors, such as prodigiosin synthesis, hemagglutination, and flagellar variation may have a common regulatory (QS) link in some strains of S. marcescens.33 These bacterial pigments have been documented to alter the immune responses and also induce cytotoxicity in the host.34,35 These pigments are also considered essential for the survival of bacteria and contribute to the pathogenicity.34 The data obtained for prodigiosin pigment inhibition is in agreement with previous findings for silver nanoparticles synthesized using Piper betle extract that reduced the prodigiosin production in S. marcescens ATCC 14756 by 72%.36
Fig. 2. (A) The inhibition of prodigiosin production and the exoprotease activity of S. marcescens MTCC 97 using AgNPs-MK. Data are represented as average values and the error bars represent the SD. The percentage inhibition is shown on the secondary y-axis. * indicates p ≤ 0.05 with respect to the control. (B) The inhibition of the swarming motility of S. marcescens MTCC 97 by AgNPs-MK: (i) control; (ii) 0.5 μg ml−1 AgNPs-MK; (iii) 4 μg ml−1 AgNPs-MK; and (iv) 8 μg ml−1 AgNPs-MK.
3.3.2. Inhibition of exoprotease activity
The proteolytic activity of S. marcescens is due to exoproteinases.37 The proteases secreted by S. marcescens cause the haemolysis of human erythrocytes.38 The protease activity in the supernatant of S. marcescens MTCC 97 was studied using an azocasein degradation assay. Significant inhibition (P-value < 0.05) of the exoprotease activity of S. marcescens MTCC 97 was observed at all tested sub-MICs (Fig. 2A). More than a 60% reduction was recorded at the highest sub-MIC (8 μg ml−1). Exoproteases are considered as one of the main virulent traits of S. marcescens. A comparative study between various strains of E. coli found that S. marcescens exhibited a more pronounced haemolysis of erythrocytes than E. coli.38 Proteases produced by S. marcescens also modulate the inflammatory responses in a host.39
3.3.3. Inhibition of swimming motility
The motility in S. marcescens is characteristic to the virulent strains and is also responsible for certain nosocomial infections such as catheter associated urinary tract infections (UTIs).24 As shown in Fig. 2B, the control S. marcescens swarmed across the entire plate within 18 h of incubation and produced a dark red pigment (prodigiosin). Treatment with 1, 2, and 4 μg ml−1 AgNPs-MK decreased the swarming motility by 25.65, 64.68, and 74.72%, respectively. More than 90% inhibition was found at 8 μg ml−1. It is also important to note that the prodigiosin biosynthesis also decreased at 8 μg ml−1 AgNPs-MK, further supporting the prodigiosin inhibition data. These flagellar-mediated motilities assist in the adherence of S. marcescens to solid surfaces, leading to the development of biofilms.40
3.4. Inhibition of the virulence factors of P. aeruginosa PAO1 by AgNPs-MK
3.4.1. Inhibition of pyocyanin production
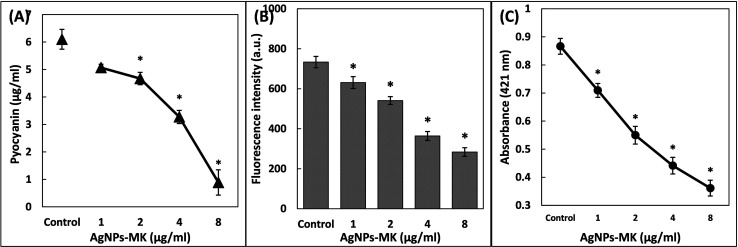
The effect of AgNPs-MK was checked against six QS-mediated virulence factors of P. aeruginosa PAO1. Pyocyanin is a blue-green pigment secreted by P. aeruginosa and is governed by bacterial cell to cell communication, for example, QS.41 The concentration of pyocyanin in the cell free supernatant of the untreated control was found to be 6.09 ± 0.36 μg ml−1. Treatment with AgNPs-MK resulted in a progressive decrease in the pigment production. The presence of 8 μg ml−1 AgNPs-MK reduced the piment by 85.44% in P. aeruginosa PAO1 as shown in Fig. 3A. Pyocyanin is known to contribute to the pathogenicity of P. aeruginosa by interfering with the cellular functions of the host's system.35 A study conducted on cystic fibrosis subjects found that pyocyanin and its precursor modulated the expression of numerous immune modulatory proteins and also caused impairment of the proper beating of human respiratory cilia.42 Pyocyanin supports the establishment of a biofilm in P. aeruginosa and suppresses the immune system of the host by inducing the apoptosis of human neutrophils.43
Fig. 3. (A) The inhibition of pyocyanin production of P. aeruginosa PAO1 using AgNPs-MK. (B) The inhibition of pyoverdin production of P. aeruginosa PAO1 using AgNPs-MK. (C) The inhibition of rhamnolipid production of P. aeruginosa PAO1 using AgNPs-MK. Data are represented as average values and the error bars represent the SD. * indicates p ≤ 0.05 with respect to the control.
3.4.2. Inhibition of pyoverdin
Pyoverdin is another pigment produced by some virulent strains of P. aeruginosa.44 The presence of 1, 2, 4, and 8 μg ml−1 AgNPs-MK inhibited pyoverdin levels in the supernatant of P. aeruginosa PAO1 by 13.95, 26.22, 50.37, and 61.30%, respectively (Fig. 3B). Pyoverdin also contributes to the virulence of P. aeruginosa. One of its mechanisms is detaching the iron form transferrin protein that ultimately results in iron deficiency in the tissues of hosts.45 Pyoverdin has been found to assist in the establishment of P. aeruginosa infections in subjects with cystic fibrosis.44 Thus, inhibition of these pigments shows the protective effect of AgNPs-MK in reducing the virulence of P. aeruginosa.
3.4.3. Inhibition of rhamnolipid production
Rhamnolipids are bacterial surfactants produced by P. aeruginosa and they play a key role in the adherence of bacterial cells to solid surfaces, as well as in the maintenance of the architecture of the biofilm.46 The production of low molecular weight rhamnolipids is directly regulated by RhlR-RhlI QS of P. aeruginosa. A subsequent reduction in rhamnolipid production was observed upon treatment with AgNPs-MK as shown in Fig. 3C. The presence of 1, 2, 4, and 8 μg ml−1 of AgNPs-MK decreased the rhamnolipid production by 18.12, 36.55, 49.05, and 58.29% in the supernatant of P. aeruginosa PAO1, respectively. The surfactant has been documented to assist in the surface motility of P. aeruginosa.47 The data clearly suggest the protective effect of AgNPs-MK on inhibiting the virulent traits of P. aeruginosa.
3.4.4. Inhibition of exoprotease activity
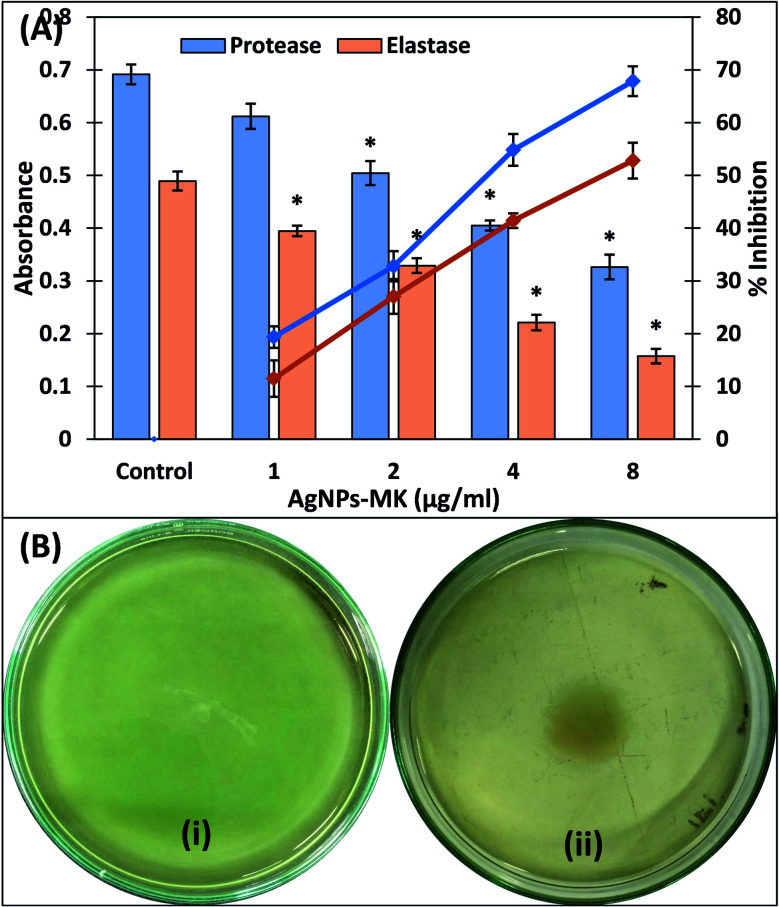
One of the major enzymes produced by virulent strains of bacteria upon successful infection are the proteolytic enzymes, these cause damage to the host's tissues. The effect of the AgNPs-MK on the inhibition of exoprotease activity was determined using a azocasein degradation assay. Treatment with 1, 2, 4, and 8 μg ml−1 AgNPs-MK resulted in an 11.47, 27.04, 41.41, and 52.79% decrease in the exoprotease activity in the supernatant of P. aeruginosa PAO1 (Fig. 4A). These proteases are secreted by pathogens and cleave the host cell's proteins, weakening the immune response and ultimately causing bacterial invasion.48 Previously, mycofabricated silver nanoparticles have been reported to inhibit the exoprotease activity of P. aeruginosa PAO1 by 86%.49
Fig. 4. (A) The inhibition of the exoprotease and elastase activities of P. aeruginosa PAO1 using AgNPs-MK. Data are represented as average values and the error bars represent the SD. The percentage inhibition is shown on the secondary y-axis. * indicates p ≤ 0.05 with respect to the control. (B) The inhibition of the swimming motility of P. aeruginosa PAO1 using AgNPs-MK: (i) control; and (ii) 8 μg ml−1 AgNPs-MK.
3.4.5. Inhibition of elastase activity
Elastases are another group of hydrolytic enzymes produced by bacteria during bacterial infection that degrade the host's tissues and suppress the immune system.50 A dose-dependent inhibition in elastinolytic activity of P. aeruginosa PAO1 was recorded in the presence of AgNPs-MK (Fig. 4A). The elastase activity was inhibited by more than 65% in the presence of the highest tested sub-MIC (8 μg ml−1) of the AgNPs-MK. The synthesis or expression of the las proteins are governed by QS in P. aeruginosa and these proteins also help the establishment of biofilms.51 A previous study reported that silver nanoparticles downregulated the lasB gene, which ultimately reduced the elastinolytic activity of the bacteria.49 Therefore, the inhibition of these enzymes (proteases and elastases) in the cell free supernatant of P. aeruginosa PAO1 signifies the modulation of lasI-lasR QS by AgNPs-MK.
3.4.6. Inhibition of swimming motility
The motility of P. aeruginosa is a key factor responsible for the spread of infection in a host and is under control of QS.52 This is considered to be an important virulence factor as it is also a key driver in the pathogenicity of P. aeruginosa.53 As shown in Fig. 4B, the control P. aeruginosa PAO1 swim across the entire Petri plate within 18 h. The zone of swimming was reduced by 35.31, 49.07, and 61.71% upon the addition of 1, 2, and 4 μg ml−1 AgNPs-MK in culture media. More than 70% inhibition was recorded at the highest sub-MIC (8 μg ml−1). The decrease in motility of P. aeruginosa PAO1 further supports and validates the anti-QS activity of the AgNPs-MK.
3.5. Inhibition of the development of biofilms by AgNPs-MK
3.5.1. Quantitative inhibition of the formation of biofilms
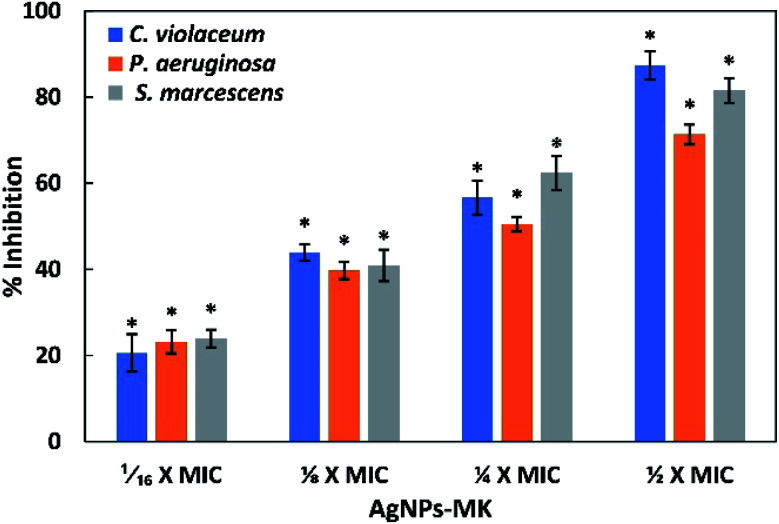
The formation of a biofilm plays a key role in the pathogenesis and it has been speculated that almost 80% of infections are established or encouraged by biofilms.14 This imposes a huge burden on the treatment of infections as the effectiveness of antibiotics is reduced up to a thousand-fold in biofilms. Often, the formation of a biofilm is regulated by an AI-mediated QS phenomenon. The effect of the synthesized AgNPs-MK on the development of biofilms was studied for all three tested bacteria (C. violaceum 12472, P. aeruginosa PAO1, and S. marcescens MTCC 97) and the results are shown Fig. 5. Treatment with 0.5, 1, 2, and 4 μg ml−1 AgNPs-MK inhibited the formation of biofilms of C. violaceum 12472 by 20.57, 43.85, 56.64, and 87.39%, respectively. More than 70% inhibition of P. aeruginosa PAO1 biofilm was found at the highest sub-MIC (8 μg ml−1). Similarly, the biofilms of S. marcescens MTCC 97 were decreased by 23.92, 40.90, 62.38, and 81.54% in the presence of 1, 2, 4, and 8 μg ml−1 of AgNPs-MK, respectively. A study has found that during the mode of growth of biofilms, bacteria not only become more resistant to chemical and physical therapeutic agents, but also coordinate the expression of their virulence genes.54 For instance, a previously published study reported that the resistance to tobramycin increased by 1000-times in P. aeruginosa in biofilms compared to their planktonic counterparts.55 Silver nanoparticles synthesized using bark extract of Holarrhena pubescens have been found to inhibit the formation of biofilms of imipenem-resistant P. aeruginosa.56 Moreover, silver nanoparticles have also been reported to inhibit the biofilm formation of EsβL, producing P. aeruginosa and methicillin-resistant S. aureus.57 These findings clearly demonstrate the broad-spectrum inhibition of biofilms of Gram negative bacteria by the AgNPs-MK.
Fig. 5. The inhibition of the formation of biofilms of C. violaceum 12472, P. aeruginosa PAO1, and S. marcescens MTCC 97 using AgNPs-MK. Data are represented as average values and the error bars represent the SD. * indicates p ≤ 0.05 with respect to the control. The concentrations 1/16 × MIC, 1/8 × MIC, 1/4 × MIC, and 1/2 × MIC of AgNPs-MK against C. violaceum 12472 are 0.5, 1, 2, and 4 μg ml−1, respectively. The concentrations 1/16 × MIC, 1/8 × MIC, 1/4 × MIC, and 1/2 × MIC of AgNPs-MK against P. aeruginosa PAO1 and S. marcescens MTCC 97 are 1, 2, 4, and 8 μg ml−1, respectively.
3.5.2. Microscopic analysis of the inhibition of biofilms on a glass surface
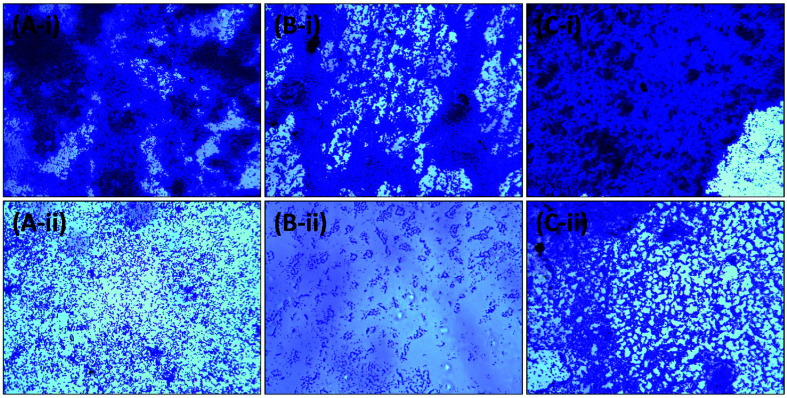
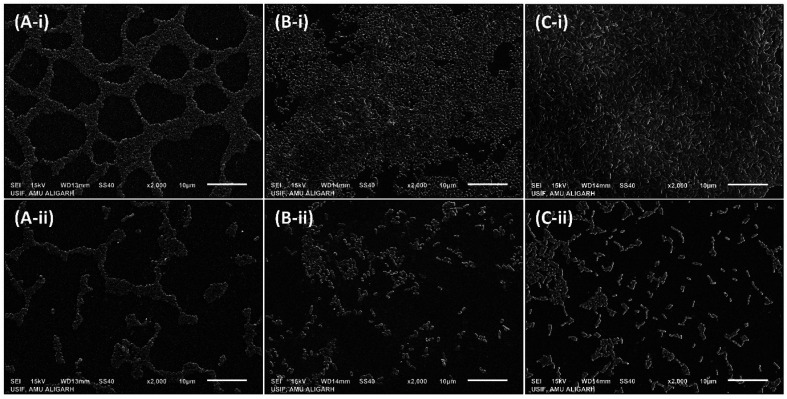
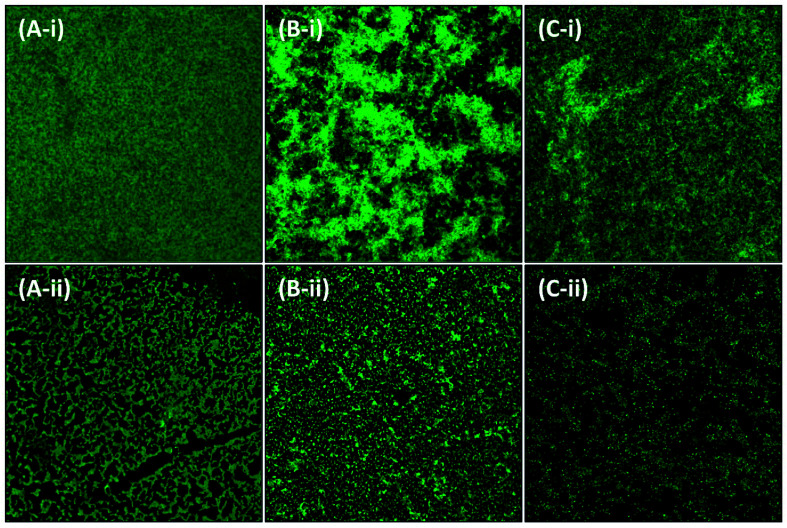
Further analysis of the inhibition of biofilms was performed using microscopy. The test bacteria were cultured in the absence and presence of the highest sub-MIC of AgNPs-MK, respectively, to visualize the changes in the architecture of the biofilm. As evident from the light microscopic images (Fig. 6), a thick cluster of cells can be seen on the glass coverslips of all tested bacteria. The treatment with AgNPs-MK remarkably reduced the clustering of cells on the glass surface and cells be seen in a scattered form. Similarly, SEM revealed that the biofilms of the control test bacteria showed extensive biofilm formation with a normal cellular morphology (Fig. 7). The bacterial cells can also be seen enclosed in a polymeric matrix (EPS) forming a clump of cells. Treatment with the respective highest sub-MICs (4 μg ml−1 for C. violaceum 12472, 8 μg ml−1 for P. aeruginosa PAO1, and 8 μg ml−1 for S. marcescens MTCC 97) reduced the colonisation and biofilm formation of the bacteria. Moreover, the aggregation of the bacterial cells was reduced and a polymeric matrix encapsulating bacterial cells was not found upon treatment with the AgNPs-MK. A similar observation was found in the confocal microscopy study (Fig. 8). The control bacteria formed dense biofilms with many layers of cells on the glass surface. The presence of the AgNPs-MK in culture media decreased the colonization of bacteria, cells were found to be scattered and a single layer of bacterial cells was seen. The microscopic analysis validates the quantitative biofilm data showing a broad-spectrum inhibition of biofilms by the synthesized AgNPs-MK.
Fig. 6. Representative micrographs obtained using light microscopy of biofilms showing the effects of AgNPs-MK at the respective highest sub-MICs: (A-i) control C. violaceum 12472; (A-ii) C. violaceum 12472 treated with 4 μg ml−1 AgNPs-MK; (B-i) control P. aeruginosa PAO1; (B-ii) P. aeruginosa PAO1 treated with 8 μg ml−1 AgNPs-MK; (C-i) control S. marcescens MTCC 97; and (C-ii) S. marcescens MTCC 97 treated with 8 μg ml−1 AgNPs-MK.
Fig. 7. Representative micrographs obtained using scanning electron microscopy of biofilms showing the effects of AgNPs-MK at the respective highest sub-MICs: (A-i) control S. marcescens MTCC 97; (A-ii) S. marcescens MTCC 97 treated with 8 μg ml−1 AgNPs-MK; (B-i) control P. aeruginosa PAO1; (B-ii) P. aeruginosa PAO1 treated with 8 μg ml−1 AgNPs-MK; (C-i) control C. violaceum 12472; and (C-ii) C. violaceum 12472 treated with 4 μg ml−1 AgNPs-MK.
Fig. 8. Representative micrographs obtained using confocal microscopy of biofilms showing the effects of AgNPs-MK at the respective highest sub-MICs: (A-i) control C. violaceum 12472; (A-ii) C. violaceum 12472 treated with 4 μg ml−1 AgNPs-MK; (B-i) control S. marcescens MTCC 97; (B-ii) S. marcescens MTCC 97 treated with 8 μg ml−1 AgNPs-MK; (C-i) control P. aeruginosa PAO1; and (C-ii) P. aeruginosa PAO1 treated with 8 μg ml−1 AgNPs-MK.
A previously published study reported that nanocrystalline silver particles significantly decreased the viability of bacterial cells within biofilms of P. aeruginosa and up to 90% of bacterial cells within biofilms were killed, even at lower doses of silver nanoparticles. The study concluded that the possible application of silver nanoparticles in dressings may improve the wound by killing the bacterial cells residing in biofilms.58 The decrease in the viability of bacterial cells in biofilms has been confirmed by double staining (propidium iodide and Con A-FITC) in which the 3D structure of the biofilm was disrupted and a decrease in the glycocalyx matrix was found by the treatment of silver nanoparticles.59 Therefore, the data clearly validated the broad-spectrum antibiofilm activity of the AgNPs-MK synthesized using M. koenigii extract against Gram negative bacterial pathogens.
4. Conclusions
Greenly synthesized AgNPs-MK remarkably reduced the multiple QS-regulated functions of Gram negative bacterial pathogens, such as P. aeruginosa PAO1, S. marcescens MTCC 97, and C. violaceum 12472. An inhibition of violacein pigment production in C. violaceum 12472 of more than 80% was recorded. The QS-controlled virulent traits of S. marcescens MTCC 97 were also reduced by up to 90% upon treatment with AgNPs-MK. The nanoparticles also inhibited the virulence factors of P. aeruginosa PAO1 in a dose-dependent manner at sub-MICs. The formation of biofilms of all test bacteria was decreased by more than 70% at their highest respective sub-MICs. Moreover, the formation of biofilms on the surfaces of glass coverslips was remarkably inhibited, and the aggregation and formation of EPS were almost not visible. In conclusion, greenly synthesized AgNPs-MK could be exploited for the treatment of topical/skin infections. Moreover, they may also be used for coating the surfaces of medical implants/devices to prevent bacterial adherence and the development of biofilms. Further in vivo assays are needed to evaluate the therapeutic efficacy against infections caused by drug resistant bacterial pathogens.
Abbreviations
- AgNPs-MK
Silver nanoparticles synthesized using M. koenigii extract
- AHL
Acyl homoserine lactone
- AI
Autoinducer
- CLSM
Confocal laser scanning microscopy
- ECR
Elastin congo red
- QS
Quorum sensing
- SEM
Scanning electron microscopy
Data availability
The data that support the findings of this study are available from the corresponding author (IA) upon reasonable request.
Conflicts of interest
The authors declare that there are no conflicts of interest. We wish to confirm that there are no known conflicts of interest associated with this publication.
Supplementary Material
Acknowledgments
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group project number RG-1439-076. FAQ is thankful to CSIR [File no. 09/112(0626)2k19 EMR] for providing SRF.
References
- Vikesland P. Garner E. Gupta S. Kang S. Maile-Moskowitz A. Zhu N. Acc. Chem. Res. 2019;52:916–924. doi: 10.1021/acs.accounts.8b00643. [DOI] [PubMed] [Google Scholar]
- WHO, The Top 10 Causes of Death, 2018 [Google Scholar]
- Shakoor S. Platts-Mills J. A. Hasan R. Infect. Dis. Clin. North Am. 2019;33:1105–1123. doi: 10.1016/j.idc.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Andleeb S., Majid M. and Sardar S., in Antibiotics and Antimicrobial Resistance Genes in the Environment, Elsevier, 2020, pp. 269–291 [Google Scholar]
- Dadgostar P. Infect. Drug Resist. 2019;12:3903–3910. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, No Time to Wait: Securing the Future from Drug-Resistant Infections, 2019 [Google Scholar]
- Ahmad I., Qais F. A., Samreen, Abulreesh H. H., Ahmad S. and Rumbaugh K. P., in Antibacterial Drug Discovery to Combat MDR, Springer Singapore, Singapore, 2019, pp. 1–21 [Google Scholar]
- Albrich W. C. Monnet D. L. Harbarth S. Emerg. Infect. Dis. 2004;10:514–517. doi: 10.3201/eid1003.030252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. J. Antimicrob. Chemother. 2002;49:925–934. doi: 10.1093/jac/dkf044. [DOI] [PubMed] [Google Scholar]
- Piewngam P. Chiou J. Chatterjee P. Otto M. Expert Rev. Anti Infect. Ther. 2020;18:499–510. doi: 10.1080/14787210.2020.1750951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. G. Parsek M. R. Pearson J. P. Iglewski B. H. Costerton J. W. Greenberg E. P. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Lopez D. Vlamakis H. Kolter R. Cold Spring Harbor Perspect. Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. Clin. Infect. Dis. 2001;33:1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- Schachter B. Nat. Biotechnol. 2003;21:361–365. doi: 10.1038/nbt0403-361. [DOI] [PubMed] [Google Scholar]
- Martins N. Rodrigues C. F. J. Clin. Med. 2020;9:722. doi: 10.3390/jcm9030722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I. del Pozo J. L. Penadés J. R. Leiva J. An. Sist. Sanit. Navar. 2005;28:163–175. doi: 10.4321/s1137-66272005000300002. [DOI] [PubMed] [Google Scholar]
- Qais F. A., Khan M. S. and Ahmad I., in Biotechnological Applications of Quorum Sensing Inhibitors, ed. V. C. Kalia, Springer Singapore, Singapore, 2018, pp. 227–244 [Google Scholar]
- Valcárcel M. López-Lorente Á. I. Trac. Trends Anal. Chem. 2016;84:1–2. doi: 10.1016/j.trac.2016.05.010. [DOI] [Google Scholar]
- Singh J. Dutta T. Kim K.-H. Rawat M. Samddar P. Kumar P. J. Nanobiotechnol. 2018;16:84. doi: 10.1186/s12951-018-0408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qais F. A. Shafiq A. Khan H. M. Husain F. M. Khan R. A. Alenazi B. Alsalme A. Ahmad I. Bioinorg. Chem. Appl. 2019;2019:4649506. doi: 10.1155/2019/4649506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloff J. Planta Med. 1998;64:711–713. doi: 10.1055/s-2006-957563. [DOI] [PubMed] [Google Scholar]
- Matz C. Deines P. Boenigk J. Arndt H. Eberl L. Kjelleberg S. Jurgens K. Appl. Environ. Microbiol. 2004;70:1593–1599. doi: 10.1128/AEM.70.3.1593-1599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater H. Crow M. Everson L. Salmond G. P. C. Mol. Microbiol. 2003;47:303–320. doi: 10.1046/j.1365-2958.2003.03295.x. [DOI] [PubMed] [Google Scholar]
- Salini R. Pandian S. K. Pathog. Dis. 2015;73:ftv038. doi: 10.1093/femspd/ftv038. [DOI] [PubMed] [Google Scholar]
- Essar D. W. Eberly L. Hadero A. Crawford I. P. J. Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbauer R. Sriyosachati S. Cox C. D. Infect. Immun. 1985;49:132–140. doi: 10.1128/iai.49.1.132-140.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain F. M. Ahmad I. Al-Thubiani A. S. Abulreesh H. H. AlHazza I. M. Aqil F. Front. Microbiol. 2017;8:727. doi: 10.3389/fmicb.2017.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E. Israel M. Landshman N. Chechick A. Blumberg S. Infect. Immun. 1982;38:716–723. doi: 10.1128/iai.38.2.716-723.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G. A. Kolter R. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Fuqua C. Parsek M. R. Greenberg E. P. Annu. Rev. Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Qais F. A. Shafiq A. Ahmad I. Husain F. M. Khan R. A. Hassan I. Microb. Pathog. 2020;144:104172. doi: 10.1016/j.micpath.2020.104172. [DOI] [PubMed] [Google Scholar]
- Wei J.-R. Lai H.-C. Int. J. Med. Microbiol. 2006;296:117–124. doi: 10.1016/j.ijmm.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Goluszko P. Nowicki B. Goluszko E. Nowicki S. Kaul A. Pham T. FEMS Microbiol. Lett. 1995;133:41–45. doi: 10.1111/j.1574-6968.1995.tb07858.x. [DOI] [PubMed] [Google Scholar]
- Liu G. Y. Nizet V. Trends Microbiol. 2009;17:406–413. doi: 10.1016/j.tim.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau G. W. Hassett D. J. Ran H. Kong F. Trends Mol. Med. 2004;10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Srinivasan R. Vigneshwari L. Rajavel T. Durgadevi R. Kannappan A. Balamurugan K. Pandima Devi K. Veera Ravi A. Environ. Sci. Pollut. Res. 2018;25:10538–10554. doi: 10.1007/s11356-017-1049-0. [DOI] [PubMed] [Google Scholar]
- Coria-Jiménez R. Zárate-Aquino C. Ponce-Ponce O. Folia Microbiol. 2004;49:321–326. doi: 10.1007/BF02931050. [DOI] [PubMed] [Google Scholar]
- Braun V. Gunther H. Neub B. Tautz C. Arch. Microbiol. 1985;141:371–376. doi: 10.1007/BF00428852. [DOI] [PubMed] [Google Scholar]
- Kida Y. Inoue H. Shimizu T. Kuwano K. Infect. Immun. 2007;75:164–174. doi: 10.1128/IAI.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagely A. Krediet C. J. Ritchie K. B. Teplitski M. ISME J. 2011;5:1609–1620. doi: 10.1038/ismej.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich L. E. P. Price-Whelan A. Petersen A. Whiteley M. Newman D. K. Mol. Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- Fothergill J. L. Panagea S. Hart C. A. Walshaw M. J. Pitt T. L. Winstanley C. BMC Microbiol. 2007;7:45. doi: 10.1186/1471-2180-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T. Kutty S. K. Tavallaie R. Ibugo A. I. Panchompoo J. Sehar S. Aldous L. Yeung A. W. S. Thomas S. R. Kumar N. Gooding J. J. Manefield M. Sci. Rep. 2015;5:8398. doi: 10.1038/srep08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek M. E. Bhatnagar A. McCarty N. A. Zughaier S. M. Interdiscip. Perspect. Infect. Dis. 2012;2012:1–10. doi: 10.1155/2012/843509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. Adams P. Infect. Immun. 1985;48:130–138. doi: 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E. Caiazza N. C. O'Toole G. A. J. Bacteriol. 2003;185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T. Curty L. K. Barja F. van Delden C. Pechère J. C. J. Bacteriol. 2000;182:5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. S. Iglewski B. H. Barbara H. Curr. Opin. Microbiol. 2003;6:56–60. doi: 10.1016/S1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- Singh B. R. Singh B. N. Singh A. Khan W. Naqvi A. H. Singh H. B. Sci. Rep. 2015;5:13719. doi: 10.1038/srep13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano P. A. Langeveld J. P. Hudson B. G. Noelken M. E. Infect. Immun. 1989;57:3783–3787. doi: 10.1128/iai.57.12.3783-3787.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. He X. Xie W. Xiong J. Sheng H. Guo S. Huang C. Zhang D. Zhang K. Can. J. Microbiol. 2014;60:227–235. doi: 10.1139/cjm-2013-0667. [DOI] [PubMed] [Google Scholar]
- Beatson S. A. Whitchurch C. B. Semmler A. B. T. Mattick J. S. J. Bacteriol. 2002;184:3598–3604. doi: 10.1128/JB.184.13.3598-3604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Yu S. Zhang Z. Wei Q. Yan L. Ai G. Liu H. Ma L. Z. Appl. Environ. Microbiol. 2014;80:6724–6732. doi: 10.1128/AEM.01237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompilio A. Crocetta V. De Nicola S. Verginelli F. Fiscarelli E. Di Bonaventura G. Front. Microbiol. 2015;6:951. doi: 10.3389/fmicb.2015.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel J. C. Ruseska I. Wright J. B. Costerton J. W. Antimicrob. Agents Chemother. 1985;27:619–624. doi: 10.1128/AAC.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S. G. Ansari M. A. Khan H. M. Jalal M. Mahdi A. A. Cameotra S. S. Bionanoscience. 2018;8:544–553. doi: 10.1007/s12668-018-0505-8. [DOI] [Google Scholar]
- Ali K. Ahmed B. Dwivedi S. Saquib Q. Al-Khedhairy A. A. Musarrat J. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0131178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostenko V. Lyczak J. Turner K. Martinuzzi R. J. Antimicrob. Agents Chemother. 2010;54:5120–5131. doi: 10.1128/AAC.00825-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M. A. Khan H. M. Khan A. A. Cameotra S. S. Alzohairy M. A. Indian J. Med. Microbiol. 2015;33:101. doi: 10.4103/0255-0857.148402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (IA) upon reasonable request.