Abstract
One of the main global concerns is the usage and spread of antibiotic resistant Salmonella serovars. The animals, humans, and environmental components interact and contribute to the rapid emergence and spread of antimicrobial resistance, directly or indirectly. Therefore, this study aimed to determine antibiotic resistance (AR) profiles of Salmonella serotypes isolated from the environment, animals, and humans in South Africa by a systematic review and meta-analysis. The preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines were followed to search four databases for studies published from 1980 to 2021, that reported the antibiotic resistance profiles of Salmonella serotypes isolated in South Africa. The AR was screened from 2930 Salmonella serotypes which were isolated from 6842 samples. The Western Cape province had high pooled prevalence estimates (PPE) of Salmonella isolates with AR profiles followed by North West, Gauteng, and Eastern Cape with 94.3%, 75.4%, 59.4%, and 46.2%, respectively. The high PPE and heterogeneity were observed from environmental samples [69.6 (95% CI: 41.7−88.3), Q = 303.643, I2 = 98.353, Q-P = 0.045], animals [41.9 (95% CI: 18.5–69.5), Q = 637.355, I2 = 98.745, Q-P = 0.577], as well as animals/environment [95.9 (95% CI: 5.4−100), Q = 55.253, I2 = 96.380, Q-P = 0.300]. The majority of the salmonella isolates were resistant to sulphonamides (92.0%), enrofloxacin and erythromycin (89.3%), oxytetracycline (77.4%), imipenem (72.6%), tetracycline (67.4%), as well as trimethoprim (52.2%), among the environment, animals, and humans. The level of multidrug-resistance recorded for Salmonella isolates was 28.5% in this review. This study has highlighted the occurrence of AR by Salmonella isolates from animals, humans, and environmental samples in South Africa and this calls for a consolidated “One Health” approach for antimicrobial resistance epidemiological research, as well as the formulation of necessary intervention measures to prevent further spread.
Keywords: antibiotic resistance, Salmonella, meta-analysis, South Africa
1. Introduction
Salmonella is a genus of Gram-negative rod-shaped bacterium of the Enterobacteriaceae family that includes more than 2700 Salmonella serotypes [1], found in three species, Salmonella bongori, Salmonella subterranean, and S. enterica [2]. Furthermore, S. enterica is divided into six subspecies namely: Indica (subsp. VI), arizonae (subsp. IIIa), houtenae (subsp. IV), enterica (subsp. I), salamae (subsp. II), and diarizonae (subsp. IIIb) [2]. Depending on the serovars and hosts, these bacterial species can cause bacteremia, typhoid fever, gastroenteritis, and non-typhoidal salmonellosis in humans and animals [3,4]. Salmonellosis is one of the most frequent and economically significant zoonotic infections of humans. In addition, it impacts all of the domestic animal species, as well as the wildlife.
Most of the Salmonella infections are limited to uncomplicated gastroenteritis that seldom need an antibiotic treatment. However, one of the main global concerns is the usage and spread of drug-resistant Salmonella serovars [3]. Different antibiotics have been used to treat Salmonella infection in animals and humans [2,5]. Humans, animals, and environmental components interact and contribute to the rapid emergence and spread of antimicrobial resistance, directly or indirectly [6]. Antimicrobial resistance (AMR) among salmonellae has evidently increased, according to surveillance data [2]. One of the causes for the emergence of AR might be due to the use of antimicrobials for metaphylaxis, prophylaxis, treatments, and growth promotion [7]. The common and unregulated use of antibiotics in medical and veterinary practice lead to a continual growth in AMR globally [6]. Regardless of their benefits, the continued release of antibiotics into the environment, as well as their potential negative impact on living organisms, is a major source of concern [8].
The development of AR in Salmonella strains is generally encoded by mutations in some chromosomal genes and plasmids acquired as a result of antibiotic selective pressure [9]. Resistance plasmids acquired due to the antibiotic selective pressure are commonly used to encode antibiotic resistance in Salmonella strains. Horizontal gene transfer plays a role in increasing the spread of AR genes. The use of ciprofloxacin is considered to be the first drug line of choice. However, ciprofloxacin-resistant isolates have been discovered in India [10,11] and Egypt [12], among other countries. This poses a challenge in the public health, especially with different AR genes found in several Salmonella serotypes, as shown in a study by El-Sharkawy et al. [12].
In the study conducted by Kalule et al. [13] in South Africa, Salmonella serovars isolated from both humans and meat samples demonstrated the existence of several multidrug resistant (MDR) strains to commonly used antibiotics, such as gentamicin and ciprofloxacin. Therefore, a greater knowledge of the possible mechanisms that lead to the emergence of AR in Salmonella species should aid in the development of more effective interventional measures, in order to decrease the spread of MDR strains between animals and humans [2].
This study includes a comprehensive evaluation of scientific literature published between January 1980 and August 2021 on AR, expressed by Salmonella serotypes, isolated from the environment, animals, and humans in South Africa.
2. Results
2.1. Overview of the Selected Studies
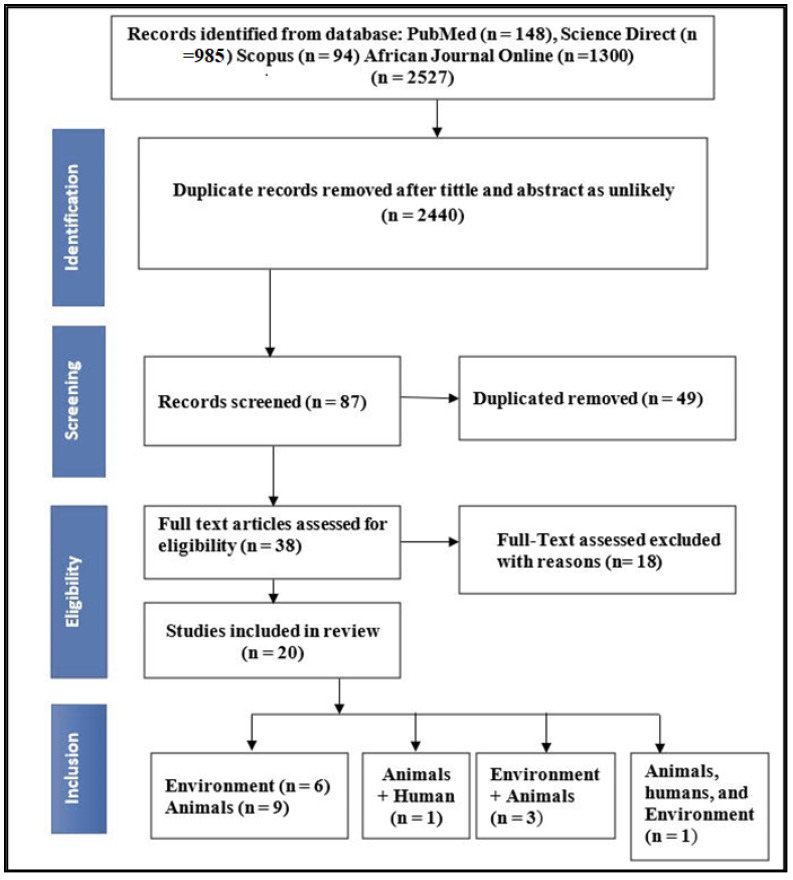
As shown in Figure 1, an electronic search of the databases PubMed, Science Direct, Africa Journals Online, and Scopus yielded 2527 articles. After screening the title and abstract, about 2440 articles were removed. Duplication resulted in the removal of 49 articles. We evaluated 38 full-text papers for eligibility, and 18 papers did not meet our requirements for various reasons: (i) Case studies (n = 5) and (ii) studies without clear total samples collected (n = 13). Finally, 20 articles with 2930 isolates were included for systemic review and meta-analysis.
Figure 1.
PRISMA flowchart illustrating the process of identifying, screening, and selecting the eligible articles used in this study.
2.2. Study Characteristics of Eligible Studies
The following are characteristics of the studies that are eligible: Articles published primarily on the quantitative prevalence of AR by Salmonella serovars/isolates in humans, animals (chicken, ducks, cows, pigs, goats, horses, and sheep), and the environment in South Africa; the type of samples and method of diagnostics used; the exact number of samples as well as the number of positives tested; articles reported in English only; and antibiotic resistance. All of the journal papers were published between the years 1980 and 2020.
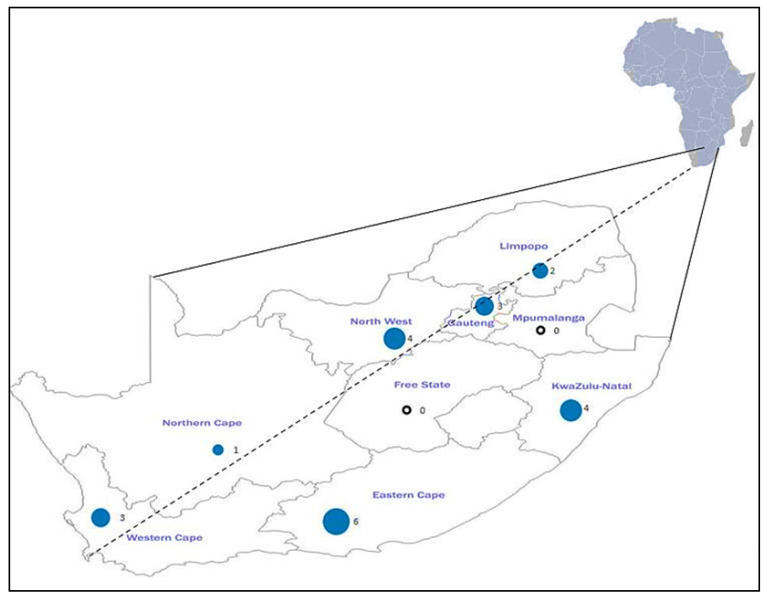
All of the studies included in this review were derived from all of the provinces in South Africa. Eastern Cape (n = 6) was observed to have the highest number of studies followed by North West (n = 4), KwaZulu-Natal (n = 4), Western Cape (n = 3), Gauteng (n = 3), Limpopo (n = 2), and Northern Cape (n = 1) as the least (Figure 2). The most common method for determining AR profiles of Salmonella serotypes isolated from all of the studies included in this systematic and meta-analysis study was disk diffusion. Of the 20 included studies, six were environmental samples, nine were samples from animal sources, and three included both animal and environmental samples. One study examined animals, humans, and environmental samples, and one study included samples from both animals and humans.
Figure 2.
Map showing the number of published studies on Salmonella antibiotic resistance per province. The black circle shows that there were no studies conducted.
2.3. Pooling and Heterogeneity Estimates of Salmonella Species
Prevalence Based on Provinces, Study Years, and Diagnostic Techniques
Table 1 displays a summary of the number of samples processed per subgroup, and the number of those samples that were positive for Salmonella serovars. Overall, the assessment of South African data revealed a pooled prevalence estimate (PPE) of 32.3% of processed samples positive for Salmonella, with a range between 41.9% and 95.9%. At 95% CI, 30.9–33.9 samples in South Africa would be expected to be positive for Salmonella serovars, and the range could be between 5.4% and 100%.
Table 1.
Pooled prevalence of Salmonella spp. from the environment, animals, and humans; screening methods; study year; and sampling sites.
| Risk Factors | Number of Studies | Pooled Estimates | Measure of Heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Size |
Number Positive | Pooled Prevalence | I2 (95% CI) | Cochran’s Q | Heterogeneity I2 (%) | Significance Level Q-p | p-Value | ||
| Overall study | |||||||||
| Environment | 6 | 1801 | 942 | 69.9 | (41.7–88.3) | 303.643 | 98.353 | 0.161 | 0.04544 |
| Animals | 9 | 3722 | 1000 | 41.9 | (18.5–69.5) | 637.355 | 98.745 | 0.577 | 0.33833 |
| Animals/humans | 1 | 200 | 146 | - | - | - | - | - | |
| Animals/environment | 3 | 904 | 834 | 95.9 | (5.4–100) | 55.253 | 96.380 | 0.304 | 0.30075 |
| Animals/humans/environment | 1 | 215 | 8 | - | - | - | - | - | |
| Study year | |||||||||
| 2000–2010 | 1 | 172 | 172 | - | - | - | - | - | |
| 2010–2021 | 19 | 6691 | 2551 | 49.8 | (33.8–65.9) | 1209.499 | 98.512 | 0.981 | 0.29987 |
| Diagnostic technique | |||||||||
| PCR | 11 | 3475 | 1551 | 60.2 | (40.9–76.8) | 608.599 | 98.357 | 0.299 | 0.46897 |
| Culture | 5 | 1463 | 778 | 76.9 | (14.6–98.5) | 89.158 | 95.514 | 0.428 | 0.16359 |
| Serotype | 3 | 1434 | 338 | 22.4 | (3.2–71.3) | 42.054 | 95.244 | 0.258 | 0.05859 |
| MALDI-TOF-MS and PCR |
1 | 481 | 263 | - | - | - | - | - | |
| Provinces | |||||||||
| KwaZulu-Natal | 4 | 2094 | 470 | 40.8 | (7.4–85.6) | 404.768 | 99.259 | 0.735 | 0.50000 |
| Gauteng | 3 | 793 | 436 | 59.4 | (4.1–98.1) | 62.831 | 96.817 | 0.832 | 0.30075 |
| Eastern Cape | 6 | 2439 | 785 | 46.2 | (21.5–73.0) | 381.864 | 98.691 | 0.796 | 0.09424 |
| North West | 4 | 169 | 528 | 75.4 | (19.4–97.5) | 250.522 | 98.802 | 0.389 | 0.24845 |
| Northern Cape | 1 | 1069 | 30 | - | - | - | - | - | |
| Limpopo | 2 | 1673 | 122 | - | - | - | - | - | |
| Western Cape | 3 | 606 | 685 | 94.3 | (1.1–100) | 75.412 | 97.348 | 0.450 | 0.30075 |
PCR = Polymerase chain reaction.
The analyses of the PPE in animals, environment, and animal/environment analyses all revealed a significant heterogeneity. For the environment, pooled heterogeneity was observed [69.6% (95% CI: 41.7–88.3), Q = 303.643, I2 = 98.353, Q-P = 0.045], animals [41.9% (95% CI: 18.5–69.5), Q = 637.355, I2 = 98.745, Q-P = 0.577], animals/environment [95.9% (95% CI: 5.4–100), Q = 55.253, I2 = 96.380, Q-P = 0.300]. Animals/humans/environment, animals/humans, and animal/humans/environment were not included on the meta-analyses due to the low number of studies.
The PPE of Salmonella isolates found in Western Cape province was 94.3% (95% CI: 1.1–100), followed by North West, Gauteng, Eastern Cape, and KwaZulu-Natal provinces with 75.4% (95% CI: 19.4–97.5); 59.4% (95% CI: 4.1–98.1); 46.2% (95% CI: 21.5–73.0); and 40.8% (95% CI: 7.4–85.6), respectively. However, Northern Cape and Limpopo were not included on the meta-analyses due to the low number of studies.
A high PPE was observed from the studies conducted during the 2010–2021 period [49.8% (95% CI: 33.8–65.9), Q = 1209.499, I2 = 98.512, Q-P = 0.981]. Studies conducted between the 2000–2010 period were not included in the meta-analysis since they were less than three.
Salmonella species were identified using three diagnostic techniques, including cultures which showed the greatest degree of sensitivity with PPE of 76.9% (95% CI: 14.6–98.5), Q = 89.158, I2 = 95.514, Q-P = 0.428, followed by PCR [60.2% (95% CI: 40.9–76.8), Q = 608.599, I2 = 98.357, Q-P = 0.299], and serotyping [22.4% (95% CI: 3.2–71.3), Q = 42.054, I2 = 95.244, Q-P = 0.258]. A combination of MALDI-TOF-MS and PCR was not included on the meta-analysis due to the low number of studies.
2.4. Antibiotic Resistance Profile of Salmonella spp. Isolates to Antibiotics
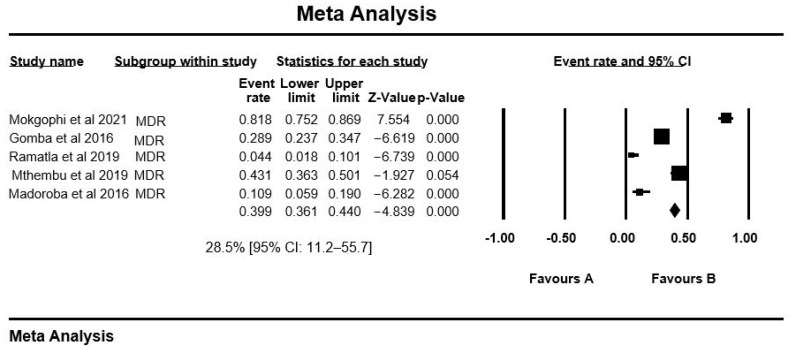
The PPE of antibiotic resistance for Salmonella serovars included in this meta-analysis (Table 1) was as follows: Sulphonamides with high PPE of 92.0% [95% CI: 37.5–99.5], enofloxacin and erythromycin 89.3% [95% CI: 62.9–97.6], oxytetracycline 77.4% [95% CI: 31.2–96.3], imipenem 72.6% [95% CI: 23.1–95.9], tetracycline 67.4% [95% CI: 53.8–78.6], trimethoprim 52.2% [95% CI: 24.7–78.4], trimethoprim-sulfamthoxazole 47.5% [95% CI: 26.3–69.6], nalidixic acid 39.8% [95% CI: 22.3–60.5], sulphamethoxazole 39.5% [95% CI: 32.6–46.8], ampicillin 38.6% [95% CI: 25.4–53.7], streptomycin 37.7% [95% CI: 17.2–63.8], and the remaining PPE of AR are shown in Table 2. Ampicillin was the most tested antibiotic with 13 studies. Overall, the MDR recorded for Salmonella spp. was 28.5% [95% CI: 11.2–55.7]. The overall effect estimates and their accompanying 95% confidence intervals (CI) did not overlap in any of the meta-analyses performed (Figure 3).
Table 2.
Pooled prevalence rate and 95% CI of antibiotic resistance of Salmonella species based on the meta-analysis.
| Antimicrobial Agents | Number of Studies | Number of Isolates | % Prevalence (95% CI) | I2 (95% CI) |
|---|---|---|---|---|
| Tetracycline | 9 | 1192 | 67.4 | (53.8–78.6) |
| Chloramphenicol | 8 | 243 | 2.6 | (14.6–28.2) |
| Ciprofloxacin | 6 | 167 | 28.9 | (8.5–63.9) |
| Sulphonamides | 3 | 285 | 92.0 | (37.5–99.5) |
| Nalidixic acid | 3 | 144 | 39.8 | (22.3–60.5) |
| Streptomycin | 9 | 593 | 37.7 | (17.2–63.8) |
| Ampicillin | 13 | 900 | 38.6 | (25.4–53.7) |
| Streptomycin | 9 | 593 | 37.7 | (17.2–63.8) |
| Amoxicillin | 3 | 80 | 19.2 | (13.8–26.1) |
| Trimethoprim | 3 | 477 | 52.2 | (24.7–78.4) |
| Enrofloxacin | 6 | 20 | 89.3 | (62.9–97.6) |
| Erythromycin | 6 | 954 | 89.3 | (62.9–97.6) |
| Gentamicin | 6 | 95 | 15.4 | (7.0–3.5) |
| Sulphamethoxazole | 3 | 165 | 39.5 | (32.6–46.8) |
| kanamycin | 6 | 172 | 26.7 | (10.1–54.1) |
| Imipenem | 3 | 517 | 72.6 | (23.1–95.9) |
| Oxytetracycline | 4 | 382 | 77.4 | (31.2–96.3) |
| Trimethoprim-sulfamthoxazole | 4 | 110 | 47.5 | (26.3–69.6) |
| MDR | 5 | 314 | 28.5 | (11.2–55.7) |
Figure 3.
Forest plot showing the pooled estimates of multidrug resistance in studies conducted in South Africa. The diamond at the base indicates the pooled estimates from the overall studies.
The AR was screened from a total of 8843 Salmonella serovars, collected from 20 studies. A total of 35 antibiotics were tested on these isolates. The majority of the isolates tested for AR vary by provinces (Table 3), which included Eastern Cape (n = 2439), followed by KwaZulu-Natal (n = 2094), Limpopo (n = 1673), Northern Cape (n = 1069), Gauteng (n = 793), Western Cape (n = 606), and finally by North West (n = 169). In terms of the antibiotic profile tested, ampicillin was the most tested antibiotic in all of the provinces. A total of five studies reported multidrug resistance, which is defined as resistance to at least three classes of antibiotics.
Table 3.
Summary of Salmonella spp. resistant isolates of the most tested antibiotics.
| Number of Resistant Isolates (%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Provinces | Tetracycline | Ciprofloxacin | Chloramphenicol | Ampicillin | Streptomycin | Gentamicin | Erythromycin | Kanamycin |
| KwaZulu-Natal | 52/435: (57.9%) | 16/195: (8.2%) | 32/146: (21.9%) | 257/371: (69.2%) | 40/346: (11.5%) | 49/146: (33.5%) | 18/146: (12.3%) | 5/263: (1.9%) |
| Gauteng | - | 1/170: (1.4%) | 41/433: (9.4%) | 4/263: (1.5%) | 141/433: (3.2%) | - | 170/170: (100%) | 75/146: (51.3%) |
| Eastern Cape | 543/1197: (45.3%) | 33/307: (10.7%) | 98/370: (26.4%) | 487/635: (76.6%) | 286/410: (69.7%) | 13/112: (11.6%) | 406/438: (92.6%) | 47/152: (30.9%) |
| North West | 78/114: (68.4%) | 125/198: (63.1%) | 58/198: (29.2%) | 277/498: (45.5%) | 73/198: (36.8%) | 9/198: (4.5%) | 360/384: (93.7%) | - |
| Northern Cape | - | - | - | 5/30: (16.6%) | - | - | - | - |
| Limpopo | - | - | - | 41/122: (33.6%) | - | - | - | 27/92: (29.3%) |
| Western Cape | - | - | - | - | - | 2/8: (25%) | - | - |
3. Discussion
In this review, the meta-analysis was used to evaluate the prevalence of AR in Salmonella serovars isolated from the environment, animals, and humans. This study examined 20 published articles from South Africa on AR in Salmonella serovars.
The majority (95%) of the studies included in this review were conducted between 2010 to 2021. Based on our meta-analysis, the PPE of AR against sulfonamide was 92%. Our findings correlate with other studies, which reported an increased development of AR against sulfonamide in Spain, Ghana, Central African Republic, Morocco, and Italy with 38.1% 72.4%, 29%, 25%, and 69%, respectively [14,15,16,17,18]. Sulfonamides resistance can arise from chromosomal dihydropteroate synthase (DHPS) mutations or from the acquisition of DHPS drug resistance genes, whose products have lower affinity for sulfonamides [19,20]. The presence of sul genes, which encode dihydropteroate synthase in a form that is not inhibited by the drug, is linked to sulfonamide resistance in Salmonella serotypes [20]. Generally, sulfonamide is encoded by three sul genes which are sul1, sul2, and sul3 [20]. The sul1 and sul2 genes are the most frequently reported genes found among sulfonamide-resistant isolates, and also found in plasmids of other Salmonella species which are still common in Gram-negative bacterial plasmids [19,20,21].
In this meta-analysis, the PPE of tetracycline was 67.4%. Our results are consistent with the findings of the studies conducted in Iran, United States, Saudi Arabia, and China, which reported an increasing development of AR against tetracycline of 66.9%, 63%, 90.71%, and 43%, respectively by Salmonella isolates [22,23,24,25]. In South Africa, over 70% of the antibiotics used in livestock production are available over the counter [26], and this contributes to the increased prevalence of AR in the country. Tetracyclines have been an important class of antibiotics in the health and production of food animals for decades, as they are used in veterinary medicine for illness prevention and control [27]. They have a broad spectrum and have good activity against Gram-positive and Gram-negative bacteria that cause acute infections [28].
Tetracyclines act as protein synthesis inhibitors by binding to the small ribosomal subunit and preventing aminoacyl-tRNA from attaching to the protein synthesis complex [22]. The complicated syntheses of several systems are capable of conferring tetracycline resistance, including efflux pumps, enzymatic inactivation, and mutations [22,27,29,30]. In South Africa, tetracyclines are the most often used or over-used antibiotics in the livestock production [31]. This is due to the fact that they are relatively affordable and widely available as over-the-counter veterinary medications [22,31,32].
According to a survey conducted by Eagar et al. [33], 16.7% and 12.4% of tetracyclines and sulphonamides were commonly used antibiotics in animals in South Africa between the years 2002 and 2004. The Stock Remedies Act of 1947 in South Africa consents tetracycline to be purchased over the counter without a prescription from a veterinarian [26].
The PPE of AR for streptomycin by Salmonella isolates was 37.7%. Our results are lower than the observations from other studies for resistance against streptomycin in Saudi Arabia, Italy, and Egypt with prevalence of 80%, 95%, 65.5%, and 20.1% [12,34,35,36]. Among the different varieties of aminoglycoside adenylyltransferase coding genes, aadA provides resistance to streptomycin in Salmonella [37,38]. Streptomycin is one of the most commonly antibiotic used by farmers and veterinarians in the country [31]. The widespread use of antibiotics in the treatment of bacterial infections in plants and animals could explain the high prevalence of streptomycin resistance [26].
The current review also revealed that most of the Salmonella isolates have high PPE for AR against erythromycin (89.3%). Similar findings were observed previously in studies from Nigeria, Saudi Arabia, Ghana, and South Africa [Gauteng] with 100%, 100%, 86.0%, and 94.9% resistance to erythromycin [31,34,39,40].
Amoxicillin and ampicillins are two of the most common antibiotics used globally to treat salmonellosis [41,42]. In this study, the PPE for AR by Salmonella isolates for amoxicillin and ampicillins were lower as compared to the other antibiotics with 19.2% and 38.6%, respectively. Our results were much higher than 0.43% [43] and 0% [44], which were recorded earlier. Antimicrobials, such as ampicillin and others, are still widely used in livestock production in South Africa for growth promotion, prophylaxis, and treatment [26], and in hospitals [43].
The antibiotic imipenem combined with cilastatin is commonly used to prevent its degradation by renal tubular dehydropeptidase. Resistance to this medicine has been reported in a number of bacterial species, adding to the worldwide burden of antibiotic resistance in humans. The PPE of imipenem 72.6% in this study was found to be much higher than the one conducted by Ng and Rivera [45], whereby none of the Salmonella enterica isolates were resistant to imipenem.
A total of five studies reported multidrug resistance for the current study. In addition, our results are in line with previous study [46] where multidrug resistance (MDR) was exhibited.
This is the first comprehensive study addressing AR for Salmonella serovars isolated from the environment, humans, and animals in South Africa. This study selected peer reviewed and high-quality papers to provide a summary of unbiased findings on the prevalence of AR in Salmonella species isolated from the environment, humans, and animals in South Africa. In addition, it reviews the current knowledge and aids in identifying areas where appropriate evidence is lacking, resulting in new research questions/topics. With respect to provinces, there are some provinces where few or no Salmonella AR studies were conducted. Between the study periods, there was a greater disparity in study outputs, from one publication in 2000–2010 to 19 articles published between 2010–2021. In terms of overall studies, only two AR studies were conducted on Salmonella infecting humans.
Despite the fact that we have systematized data on the occurrence of AR of Salmonella species, this study has the following limitations: The PPE of animals/humans and animals/humans/environment were not calculated since there is a single published article on each. The number of studies from some provinces were unusually high, which may have influenced the overall estimate. Moreover, few reports for Salmonella AR from humans were observed, whereby only two studies were undertaken in two provinces, namely, Western Cape and KwaZulu-Natal. Furthermore, the AR genes were not included from this meta-analysis due to the few number of studies conducted.
4. Materials and Methods
4.1. Study Design
The current systematic review and meta-analysis were conducted in accordance with the preferred reporting items for systematic reviews and meta-analyses (PRISMA 2020) guidelines for selection criteria, literature search, statistical analysis, and data extraction [47].
4.2. Search Strategy
The literature search was performed on four databases, namely, (https://www.ncbi.nlm.nih.gov/pubmed, accessed on 27–28 May 2021), ScienceDirect (https://www.sciencedirect.com, accessed on 5–7 June 2021), African Journal Online (https://www.ajol.info/index.php/ajol, accessed on 11–12 June 2021), and Scopus (https://www.scopus.com, accessed on 22–24 June 2021) from June to August 2021. The following keywords were used to search for the articles: Antibiotic resistance AND Antibiotic AND drug resistance AND bacteria resistance AND multi-drug resistance AND Salmonella species AND Human OR animal AND Environment AND salmonellosis AND South Africa. We conducted our last search on 24 June 2021.
4.3. Inclusion and Exclusion Criteria
The following inclusion criteria had to be met for all of the eligible studies included in the review: Articles published primarily on the quantitative prevalence of Salmonella spp. in the environment, animals, and humans in South Africa; the type of samples and method of diagnostics used; the exact number of samples as well as the number of positives tested; and articles reported in English only on antibiotic resistance, published between January 1980 and August 2021. The studies were excluded if they were not undertaken in South Africa (Supplementary Materials). In addition, books and book chapters were excluded. Moreover, a review, a smaller sample size, and articles not reported in English, were discarded, as well as articles not published between January 1980 and August 2021.
4.4. Data extraction and Data Collection
The titles and abstracts of journal articles were examined and downloaded, and the full versions of potentially relevant articles were obtained to determine eligibility. Data including names of authors, publication year, location, total sample size, and standard methods to detect the antibiotic resistant were collected from each publication independently. Then, they were entered into a spreadsheet (Microsoft Excel®), tables, and a word document template. Only Salmonella species/isolates/serotypes specific journal articles were included in the meta-analysis.
The studies with insufficient data were excluded. In addition, review articles and studies with an abstract, but without a full text, were excluded. The papers that included the first author, year of publication, year of study, sampling, region, sample size, target population, method, drug susceptibility test, sample type, number of isolates, and number of multidrug resistance strains were considered and included in this study. For inclusion in the study, we considered all of the standard guidelines, although only the clinical and laboratory standards institute (CLSI) guidelines were used in all of the included studies (Table 4).
Table 4.
Studies included in this review, as well as provinces, methods, isolation sources, and Salmonella spp./isolates that were tested for antibiotic resistance.
| Study (Citation) | Province | Method | Sample Size | No. of Isolates | Isolate Source | Salmonella spp. |
|---|---|---|---|---|---|---|
| Gouws et al., 2000 [48] | Western Cape | DDM | Culture | 442 | Animal/environment | Salmonella spp. |
| Mokgophi et al., 2021 [31] | Gauteng | DDM | PCR | 170 | Animal | S. Bovismorbificans (58.5%); S. Dublin (18.5%); S. Enteritidis (15.7%); S. Mbandaka (12.8%); S. Saintpaul (8.5%); S. Thompson (2.8%); S. Infantis (2.8%); and S. Agona (1.4%). |
| Gomba et al., 2016 [49] | Gauteng | DDM | MALTI-TOF-MS and PCR | 263 | Environment | S. Muenchen (33.3%); S. Typhimurium (12/39; 30.8%); S. Heidelberg (20.5%); S. Bsilla (7.7%). |
| Ramatla et al., 2019 [50] | North West | DDM | PCR | 114 | Animal | S. Typhimurium (n = 44, 30.5%); S. Enteritidis (n = 18, 12.5%); S. newport (7.6%); S. Heidelberg (11.1%); S. bongori (9%); S. enterica serovar Paratyphi B (4.8%); S. Tennessee (2%); and S. Pullorum (1.3%). |
| Adesiyun et al., 2020 [51] | Gauteng | DDM | Culture | 3 | Animal | Salmonella spp. |
| Mafu et al., 2012 [52] | Eastern Cape | DDM | Culture | 40 | Environment | Salmonella spp. |
| Jaja et al., 2019 [26] | Eastern Cape | DDM | PCR | 112 | Animal | S. Enteritidis |
| Mthembu et al., 2019 [53] | Eastern Cape and KwaZulu-Natal | DDM | PCR | 194 | Environment | Salmonella spp. |
| Iwu et al., 2016 [54] | Eastern Cape | DDM | PCR | 258 | Animal | Salmonella spp. |
| Akinola et al., 2019 [55] | North West | DDM | PCR | 84 | Animal | S. bongori (10.09%); S. Pullorum (1.81%); S. Typhimurium (12.72%); S. Weltevreden; S. Chingola; S. Houten; and S. Bareily (1.81%). |
| Mathole et al., 2017 [56] | Limpopo, Eastern Cape, Northern Cape, North West, and KwaZulu Natal | DDM | Serotyping | 30 | Animal | S. Chester (3.3%); S. Cardoner (3.3%); S. Sambrae (3.3%); S. Typhimurium (3.3%); S. Schwarzengrund (6.6%); S. Aarhus (3.3%); S. Pomona (33%); S. Senftenberg (3.3%); and S. Techimani (30%); unclassified Salmonella (20%). |
| Igbinosa 2015 [57] | Eastern Cape | DMD | PCR | 150 | Environment | Salmonella spp. |
| Odjadjare and Olaniran 2015 [58] | KwaZulu Natal | DMD | PCR | 200 | Environment | Salmonella spp. |
| Zishiri et al., 2016 [42] | KwaZulu Natal | DMD | PCR | 146 | Animal/human | Salmonella spp. |
| Madoroba et al., 2016 [59] | Limpopo | DDM | PCR | 92 | Animal/environment | S. Heidelberg (2.2%); S. Aberdeen (1.1%); S. Hayindongo (1.1%); S. Mbandaka (2.2%); S. Anatum (2.2%); S. Othmarschen (1.1%); S. Nigeria (2.2%); S. Tennessee (1.1%); S. Cardoner (1.1%); S. Senftenberg (2.2%); and S. Pretoria (2.2%). |
| More et al., 2017 [60] | Western Cape | DDM | Culture | 235 | Animal | Salmonella spp. |
| Kennedy et al., 2020 [61] | KwaZulu Natal | DDM | PCR | 94 | Environment | Salmonella spp. |
| Dlamini et al., 2018 [62] | North West | DDM | Serotyping | 300 | Animal/environment | Salmonella spp. |
| Kalule et al., 2019 [13] | Western Cape | DDM | Serotyping | 8 | Human/animal/environment | S. enterica |
| Chipangura et al., 2017 [63] | South Africa | DDM | Culture | 58 | Animal | Salmonella spp. |
DDM: Disc-diffusion method; PCR = polymerase chain reaction.
4.5. Data Analysis and Assessment of Risk of Bias
The Begg’s and Egger’s tests were used to investigate the possibility of propagation bias. For each study, the prevalence, effect size, and 95% CIs were calculated as a point estimate.
A comprehensive meta-analysis was used for all of the statistical analyses, version 3.0 (CMA) program (https://www.meta-analysis.com/, accessed on 18 August 2021). The software was used to generate the pooled estimates, Cochran’s Q, p-values, and forest plots. I2 (level of inconsistency) was used to assess the study heterogeneity (Cochran’s Q). The I2 values above 75% were regarded to have a high degree of heterogeneity [64]. For each study, the prevalence, effect size, and 95% CIs were calculated as a point estimate. Exploring the funnel plots was used to assess the publication bias.
5. Conclusions
This systematic review and meta-analysis provided an overview of published Salmonella serovar AR scientific data from humans, animals, and environmental samples in South Africa. The higher PPE rate of Salmonella isolates were tested. In addition, expressing AR was observed in the Western Cape province. Disk diffusion was the most prevalent method for identifying the antibiotic resistance profiles of Salmonella serotypes isolated from all of the studies. The MDR has been identified as a serious and growing issue, according to the findings of this study. Our results revealed the highest PPE of Salmonella AR to sulphonamides, followed by enrofloxacin and erythromycin. In addition, this finding calls for the restricted usage of this group of antibiotics. According to our data analysis, the most tested antibiotics against Salmonella isolates are tetracycline, ciprofloxacin, chloramphenicol, ampicillin, streptomycin, gentamicin, erythromycin, and kanamycin. This study highlights the lack of published Salmonella serovar AR scientific data from humans. The development of AR to commonly prescribed antibiotics is very common, whilst it appears that surveillance has a lot of gaps whereby there has been no studies on AR in two provinces, namely, Free State and Mpumalanga. To stop the spread of Salmonella AR, control strategies should be strengthened. To describe the epidemiology of the serotypes across the country, large-scale investigations are required. Control practices should be strengthened to slow the spread of AR in South Africa and there is a need for a “One Health” collaborative research between the animal and human health sector, as well as the environmental sector on the epidemiology of AR development and necessary interventions.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10121435/s1, Table S1: Checklist of items to include when reporting a systematic review or meta-analysis, Figure S1: Forest plot of the meta-analysis of overall studies on the prevalence of Salmonella isolates in South Africa.
Author Contributions
Conceptualization, T.R. and O.T.; methodology, T.R. and M.T.; validation, T.R., K.E.L., T.E.O. and O.T.; formal analysis, T.R. and O.T.; investigation, T.R., M.T., T.E.O., K.E.L. and O.T.; writing—original draft preparation, T.R. and O.T.; writing—review and editing, T.E.O., K.E.L. and O.T.; visualization, K.E.L., T.E.O. and O.T.; supervision, T.E.O. and O.T.; project administration, T.R., M.T. and T.E.O. All authors have read and agreed to the published version of the manuscript.
Funding
This review received no external funding.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ketema L., Ketema Z., Kiflu B., Alemayehu H., Terefe Y., Ibrahim M., Eguale T. Prevalence and antimicrobial susceptibility profile of Salmonella serovars isolated from slaughtered cattle in Addis Ababa, Ethiopia. Biomed. Res. Int. 2018;2018:9794869. doi: 10.1155/2018/9794869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H.M., Wang Y., Su L.H., Chiu C.H. Nontyphoid Salmonella infection: Microbiology, clinical features, and antimicrobial therapy. Pediatr. Neonatol. 2013;54:147–152. doi: 10.1016/j.pedneo.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Mengistu G., Dejenu G., Tesema C., Arega B., Awoke T., Alemu K., Moges F. Epidemiology of streptomycin resistant Salmonella from humans and animals in Ethiopia: A systematic review and meta-analysis. PLoS ONE. 2020;15:e0244057. doi: 10.1371/journal.pone.0244057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernal-Bayard J., Ramos-Morales F. Molecular mechanisms used by Salmonella to evade the immune system. Curr. Issues Mol. Biol. 2018;25:133–168. doi: 10.21775/cimb.025.133. [DOI] [PubMed] [Google Scholar]
- 5.Gordon D. Ceftriaxone-resistant Salmonella infection from antibiotic-treated cattle. Gastroenterology. 2000;119:3–4. doi: 10.1016/S0016-5085(00)70050-3. [DOI] [PubMed] [Google Scholar]
- 6.Ateba C.N., Tabi N.M., Fri J., Bissong M.E.A., Bezuidenhout C.C. Occurrence of Antibiotic-Resistant Bacteria and Genes in Two Drinking Water Treatment and Distribution Systems in the North-West Province of South Africa. Antibiotics. 2020;9:745. doi: 10.3390/antibiotics9110745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaja I.F., Bhembe N.L., Green E., Oguttu J., Muchenje V. Molecular characterisation of antibiotic-resistant Salmonella enterica isolates recovered from meat in South Africa. Acta Trop. 2019;190:129–136. doi: 10.1016/j.actatropica.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Cycoń M., Mrozik A., Piotrowska-Seget Z. Antibiotics in the soil environment—Degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019;10:338. doi: 10.3389/fmicb.2019.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carattoli A. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 2003;5:113–122. [PubMed] [Google Scholar]
- 10.Gokul B.N., Menezes G.A., Harish B.N. Acc-1 β-lactamase–producing Salmonella enterica serovar Typhi, India. Emerg. Infect. Dis. 2010;16:1170–1171. doi: 10.3201/eid1607.091643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menezes G.A., Khan M.A., Harish B.N., Parija S.C., Goessens W., Vidyalakshmi K., Baliga S., Hays J. Molecular characterization of antimicrobial resistance in non-typhoidal Salmonellae associated with systemic manifestations from India. J. Med. Microbiol. 2010;59:1477–1483. doi: 10.1099/jmm.0.022319-0. [DOI] [PubMed] [Google Scholar]
- 12.El-Sharkawy H., Tahoun A., El-Gohary A.E.G.A., El-Abasy M., El-Khayat F., Gillespie T., Kitade Y., Hafez H.M., Neubauer H., El-Adawy H. Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathog. 2017;9:1–12. doi: 10.1186/s13099-017-0157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalule J.B., Smith A.M., Vulindhlu M., Tau N.P., Nicol M.P., Keddy K.H., Robberts L. Prevalence and antibiotic susceptibility patterns of enteric bacterial pathogens in human and non-human sources in an urban informal settlement in Cape Town, South Africa. BMC Microbiol. 2019;19:1–11. doi: 10.1186/s12866-019-1620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Márquez F.M.L., Burgos M.J.G., Pulido R.P., Gálvez A., López R.L. Biocide tolerance and antibiotic resistance in Salmonella isolates from hen eggshells. Foodborne Pathog. Dis. 2017;14:89–95. doi: 10.1089/fpd.2016.2182. [DOI] [PubMed] [Google Scholar]
- 15.Agyare C., Boamah V.E., Zumbi C.N., Osei F.B. Antibiotic use in poultry production and its effects on bacterial resistance. In: Kumar Y., editor. Antibiotic Resistance—A Global Threat. IntechOpen; London, UK: 2019. [Google Scholar]
- 16.Mossoro-Kpinde C.D., Manirakiza A., Mbecko J.R., Misatou P., Le Faou A., Frank T. Antimicrobial resistance of enteric Salmonella in Bangui, Central African Republic. J. Trop. Med. 2015;2015:483974. doi: 10.1155/2015/483974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amajoud N., Bouchrif B., El Maadoudi M., Senhaji N.S., Karraouan B., El Harsal A., El Abrini J. Prevalence, serotype distribution, and antimicrobial resistance of Salmonella isolated from food products in Morocco. J. Infect. Dev. Ctries. 2017;11:136–142. doi: 10.3855/jidc.8026. [DOI] [PubMed] [Google Scholar]
- 18.Proroga Y.T., Capuano F., Carullo M.R., La Tela I., Capparelli R., Barco L., Pasquale V. Occurrence and antimicrobial resistance of Salmonella strains from food of animal origin in southern Italy. Folia Microbiol. 2016;61:21–27. doi: 10.1007/s12223-015-0407-x. [DOI] [PubMed] [Google Scholar]
- 19.Guerra B., Junker E., Helmuth R. Incidence of the recently described sulfonamide resistance gene sul3 among German Salmonella enterica strains isolated from livestock and food. Antimicrob. Agents Chemother. 2004;48:2712–2715. doi: 10.1128/AAC.48.7.2712-2715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mąka Ł., Maćkiw E., Ścieżyńska H., Modzelewska M., Popowska M. Resistance to sulfonamides and dissemination of sul genes among Salmonella spp. isolated from food in Poland. Foodborne Pathog. Dis. 2015;12:383–389. doi: 10.1089/fpd.2014.1825. [DOI] [PubMed] [Google Scholar]
- 21.Infante B., Grape M., Larsson M., Kristiansson C., Pallecchi L., Rossolini G.M., Kronvall G. Acquired sulphonamide resistance genes in faecal Escherichia coli from healthy children in Bolivia and Peru. Int. J. Antimicrob. Agents. 2005;25:308–312. doi: 10.1016/j.ijantimicag.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Vaez H., Ghanbari F., Sahebkar A., Khademi F. Antibiotic resistance profiles of Salmonella serotypes isolated from animals in Iran: A meta-analysis. Iran. J. Vet. Res. 2020;21:188–197. [PMC free article] [PubMed] [Google Scholar]
- 23.Velasquez C.G., Macklin K.S., Kumar S., Bailey M., Ebner P.E., Oliver H.F., Martin-Gonzalez F.S., Singh M. Prevalence and antimicrobial resistance patterns of Salmonella isolated from poultry farms in southeastern United States. Poult. Sci. 2018;97:2144–2152. doi: 10.3382/ps/pex449. [DOI] [PubMed] [Google Scholar]
- 24.Elhadi N. Prevalence and antimicrobial resistance of Salmonella spp. in raw retail frozen imported freshwater fish to Eastern Province of Saudi Arabia. Asian Pac. J. Trop. Biomed. 2014;4:234–238. doi: 10.1016/S2221-1691(14)60237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Biswas S., Paudyal N., Pan H., Li X., Fang W., Yue M. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 2019;10:985. doi: 10.3389/fmicb.2019.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaja I.F., Oguttu J., Jaja C.J.I., Green E. Prevalence and distribution of antimicrobial resistance determinants of Escherichia coli isolates obtained from meat in South Africa. PLoS ONE. 2020;15:e0216914. doi: 10.1371/journal.pone.0216914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van T.T.H., Yidana Z., Smooker P.M., Coloe P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020;20:170–177. doi: 10.1016/j.jgar.2019.07.031. [DOI] [PubMed] [Google Scholar]
- 28.Ramatla T.A. A Study of Antibiotic Residues in Meat Sold in Butcheries and Supermarkets Around Mafikeng, North West Province (Doctoral Dissertation, North-West University (South Africa) 2016. [(accessed on 12 July 2021)]. Available online: https://repository.nwu.ac.za/bitstream/handle/10394/35485/Ramatla_TA.pdf?sequence=1.
- 29.Hao H., Sander P., Iqbal Z., Wang Y., Cheng G., Yuan Z. The risk of some veterinary antimicrobial agents on public health associated with antimicrobial resistance and their molecular basis. Front. Microbiol. 2016;7:1626. doi: 10.3389/fmicb.2016.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miruka O.D., Rose K., Nyandago W.E. Tetracycline efflux pump in different Salmonella enterica isolated from diarrhea patients in two rural health centers in Western Kenya. Iran. J. Clin. Infect. Dis. 2011;6:24–30. [Google Scholar]
- 31.Mokgophi T.M., Gcebe N., Fasina F., Adesiyun A.A. Antimicrobial resistance profiles of Salmonella isolates on chickens processed and retailed at outlets of the informal market in Gauteng Province, South Africa. Pathogens. 2021;10:273. doi: 10.3390/pathogens10030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theobald S., Etter E.M.C., Gerber D., Abolnik C. Antimicrobial resistance trends in Escherichia coli in South African poultry: 2009–2015. Foodborne Pathog. Dis. 2019;16:652–660. doi: 10.1089/fpd.2018.2612. [DOI] [PubMed] [Google Scholar]
- 33.Eagar H., Swan G., Van Vuuren M. A survey of antimicrobial usage in animals in South Africa with specific reference to food animals. J. S. Afr. Vet. Assoc. 2012;83:1–8. doi: 10.4102/jsava.v83i1.16. [DOI] [PubMed] [Google Scholar]
- 34.El-Tayeb M.A., Ibrahim A.S., Al-Salamah A.A., Almaary K.S., Elbadawi Y.B. Prevalence, serotyping and antimicrobials resistance mechanism of Salmonella enterica isolated from clinical and environmental samples in Saudi Arabia. Braz. J. Microbiol. 2017;48:499–508. doi: 10.1016/j.bjm.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pezzella C., Ricci A., DiGiannatale E., Luzzi I., Carattoli A. Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob. Agents Chemother. 2004;48:903–908. doi: 10.1128/AAC.48.3.903-908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun T., Liu Y., Qin X., Aspridou Z., Zheng J., Wang X., Li Z., Dong Q. The Prevalence and Epidemiology of Salmonella in Retail Raw Poultry Meat in China: A Systematic Review and Meta-Analysis. Foods. 2021;10:2757. doi: 10.3390/foods10112757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcaine S.D., Warnick L.D., Wiedmann M. Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 2007;70:780–790. doi: 10.4315/0362-028X-70.3.780. [DOI] [PubMed] [Google Scholar]
- 38.Nair D.V.T., Venkitanarayanan K., Johny A.K. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods. 2018;7:167. doi: 10.3390/foods7100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beshiru A., Igbinosa I.H., Igbinosa E.O. Prevalence of antimicrobial resistance and virulence gene elements of Salmonella serovars from ready-to-eat (RTE) shrimps. Front. Microbiol. 2019;10:1613. doi: 10.3389/fmicb.2019.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adzitey F., Asiamah P., Boateng E.F. Prevalence and antibiotic susceptibility of Salmonella enterica isolated from cow milk, milk products and hands of sellers in the Tamale Metropolis of Ghana. J. Appl. Sci. Environ. Manag. 2020;24:59–64. doi: 10.4314/jasem.v24i1.8. [DOI] [Google Scholar]
- 41.de Toro M., Sáenz Y., Cercenado E., Rojo-Bezares B., García-Campello M., Undabeitia E., Torres C. Genetic characterization of the mechanisms of resistance to amoxicillin/clavulanate and third-generation cephalosporins in Salmonella enterica from three Spanish hospitals. Int. Microbiol. 2011;14:173–181. doi: 10.2436/20.1501.01.146. [DOI] [PubMed] [Google Scholar]
- 42.Zishiri O.T., Mkhize N., Mukaratirwa S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J. Vet. Res. 2016;83:1–11. doi: 10.4102/ojvr.v83i1.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen W., Chen H., Geng J., Wu R.A., Wang X., Ding T. Prevalence, serovar distribution, and antibiotic resistance of Salmonella spp. isolated from pork in China: A systematic review and meta-analysis. Int. J. Food Microbiol. 2021;361:109473. doi: 10.1016/j.ijfoodmicro.2021.109473. [DOI] [PubMed] [Google Scholar]
- 44.Matsui K., Nakazawa C., Khin S.T.M.M., Iwabuchi E., Asai T., Ishihara K. Molecular Characteristics and Antimicrobial Resistance of Salmonella enterica Serovar Schwarzengrund from Chicken Meat in Japan. Antibiotics. 2021;10:1336. doi: 10.3390/antibiotics10111336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng K.C., Rivera W.L. Antimicrobial resistance of Salmonella enterica isolates from tonsil and jejunum with lymph node tissues of slaughtered swine in Metro Manila, Philippines. Int. Sch. Res. Not. 2014;2014:364265. doi: 10.1155/2014/364265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tawyabur M., Islam M.S., Sobur M.A., Hossain M.J., Mahmud M.M., Paul S., Hossain M.T., Ashour H.M., Rahman M.T. Isolation and Characterization of Multidrug-Resistant Escherichia Coli and Salmonella Spp. from Healthy and Diseased Turkeys. Antibiotics. 2020;9:770. doi: 10.3390/antibiotics9110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page M.J., Moher D., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. PRISMA 2020 Explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ. 2021;29:372. doi: 10.1136/bmj.n160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gouws P.A., Brozel V. Antimicrobial resistance of Salmonella isolates associated with retail chicken and a poultry abattoir. S. Afr. J. Sci. 2000;96:254–256. [Google Scholar]
- 49.Gomba A., Chidamba L., Korsten L. Antimicrobial resistance profiles of Salmonella spp. from agricultural environments in fruit production systems. Foodborne Pathog. Dis. 2016;13:495–501. doi: 10.1089/fpd.2016.2120. [DOI] [PubMed] [Google Scholar]
- 50.Ramatla T., Taioe M.O., Thekisoe O.M., Syakalima M. Confirmation of antimicrobial resistance by using resistance genes of isolated Salmonella spp. in chicken houses of North West, South Africa. World Vet. J. 2019;9:158–165. doi: 10.36380/scil.2019.wvj20. [DOI] [Google Scholar]
- 51.Adesiyun A.A., Nkuna C., Mokgoatlheng-Mamogobo M., Malepe K., Simanda L. Food safety risk posed to consumers of table eggs from layer farms in Gauteng Province, South Africa: Prevalence of Salmonella species and Escherichia coli, antimicrobial residues, and antimicrobial resistant bacteria. J. Food Saf. 2020;40:e12783. doi: 10.1111/jfs.12783. [DOI] [Google Scholar]
- 52.Mafu N.C., Pironcheva G., Okoh A.I. Genetic diversity and in vitro antibiotic susceptibility profile of Salmonella species isolated from domestic water and wastewater sources in the Eastern Cape Province of South Africa. Afr. J. Biotechno. 2009;8:1263–1269. [Google Scholar]
- 53.Mthembu T.P., Zishiri O.T., El Zowalaty M.E. Molecular detection of multidrug-resistant Salmonella isolated from livestock production systems in South Africa. Infect. Drug Resist. 2019;12:3537–3548. doi: 10.2147/IDR.S211618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwu C.J., Iweriebor B.C., Obi L.C., Basson A.K., Okoh A.I. Multidrug-resistant Salmonella isolates from swine in the Eastern Cape province, South Africa. J. Food Prot. 2016;79:1234–1239. doi: 10.4315/0362-028X.JFP-15-224. [DOI] [PubMed] [Google Scholar]
- 55.Akinola S.A., Mwanza M., Ateba C.N. Occurrence, genetic diversities and antibiotic resistance profiles of Salmonella serovars isolated from chickens. Infect. Drug Resist. 2019;12:3327–3342. doi: 10.2147/IDR.S217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathole M.A., Muchadeyi F.C., Mdladla K., Malatji D.P., Dzomba E.F., Madoroba E. Presence, distribution, serotypes and antimicrobial resistance profiles of Salmonella among pigs, chickens and goats in South Africa. Food Control. 2017;72:219–224. doi: 10.1016/j.foodcont.2016.05.006. [DOI] [Google Scholar]
- 57.Igbinosa I.H. Prevalence and detection of antibiotic-resistant determinant in Salmonella isolated from food-producing animals. Trop. Anim. Health Prod. 2015;47:37–43. doi: 10.1007/s11250-014-0680-8. [DOI] [PubMed] [Google Scholar]
- 58.Odjadjare E.C., Olaniran A.O. Prevalence of antimicrobial resistant and virulent Salmonella spp. in treated effluent and receiving aquatic milieu of wastewater treatment plants in Durban, South Africa. Int. J. Environ. Res. Public Health. 2015;12:9692–9713. doi: 10.3390/ijerph120809692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madoroba E., Gelaw A.K., Kapeta D. Salmonella contamination, serovars and antimicrobial resistance profiles of cattle slaughtered in South Africa. Onderstepoort J. Vet. Res. 2016;83:1–8. doi: 10.4102/ojvr.v83i1.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moré E., Ayats T., Ryan P.G., Naicker P.R., Keddy K.H., Gaglio D., Witteveen M., Cerdà-Cuéllar M. Seabirds (Laridae) as a source of Campylobacter spp., Salmonella spp. and antimicrobial resistance in South Africa. Environ. Microbiol. 2017;19:4164–4176. doi: 10.1111/1462-2920.13874. [DOI] [PubMed] [Google Scholar]
- 61.Kennedy B., Shobo C.O., Zishiri O.T., Bester L.A. Surveillance of Salmonella spp. in the environment of public hospitals in KwaZulu-Natal, South Africa. J. Hosp. Infect. 2020;105:205–212. doi: 10.1016/j.jhin.2020.02.019. [DOI] [PubMed] [Google Scholar]
- 62.Dlamini B.S., Montso P.K., Kumar A., Ateba C.N. Distribution of virulence factors, determinants of antibiotic resistance and molecular fingerprinting of Salmonella species isolated from cattle and beef samples: Suggestive evidence of animal-to-meat contamination. Environ. Sci. Pollut. Res. Int. 2018;25:32694–32708. doi: 10.1007/s11356-018-3231-4. [DOI] [PubMed] [Google Scholar]
- 63.Chipangura J.K., Chetty T., Kgoete M., Naidoo V. Prevalence of antimicrobial resistance from bacterial culture and susceptibility records from horse samples in South Africa. Prev. Vet. Med. 2017;148:37–43. doi: 10.1016/j.prevetmed.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 64.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.