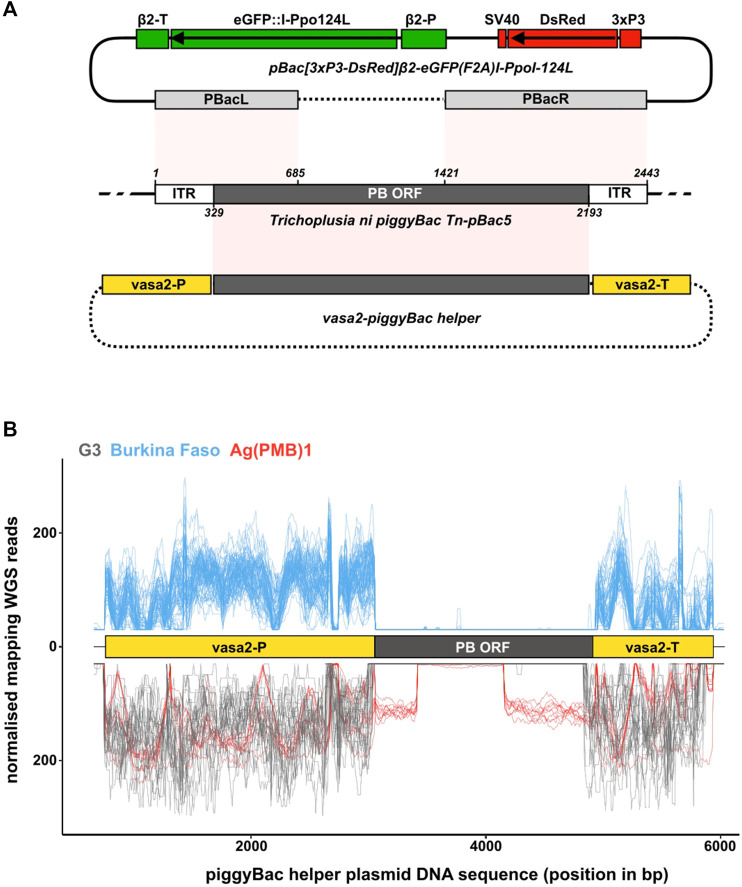
FIGURE 1.
piggyBac transposase components in laboratory and wild type individuals. (A) Schematic of a wild type piggyBac (PB) transposon from Trichoplusia ni (middle; NCBI accession DQ236240.1), the Ag(PMB)1 transformation construct [(Galizi et al., 2014); top] and the PB helper plasmid [(Volohonsky et al., 2015); bottom]. Shown are the regions of the endogenous PB locus present in the two microinjected plasmids highlighting how both the transformation and helper plasmid lack the complete machinery required for transposon mobility. pBacL (left) and pBacR (right) arms present in the pBac[3xP3-DsRed]β2-eGFP:I-PpoI-124L transformation construct contain the entire flanking inverted terminal repeats (ITRs) and partial regions of the PB open reading frame (ORF). The helper PB plasmid containing the complete PB ORF driven from the vasa2 regulatory regions lacks the flanking ITRs. Sequences of the transformation construct that are integrated in the genome, and those present only transiently in injected individuals are shown in solid lines and dashed lines, respectively. (B) Mapping of whole genome sequencing reads from the G3 and Ag(PMB)1 controls (bottom in negative y-axis; grey and red) and the 81 wild type individuals collected in Burkina Faso villages (top; blue). The position of the An. gambiae vasa regulatory regions (yellow boxes) and the PB transposase ORF (black box) is also shown. Reads are normalized by scaling counts to the number of reads in the most abundant sample.