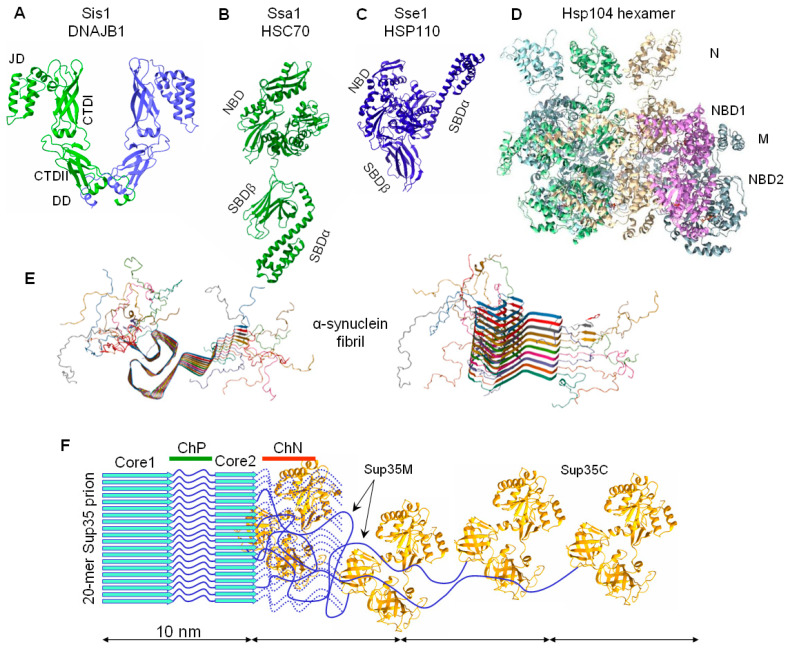
Figure 3.
Relative sizes of a α-synuclein fibril, Sup35 prion, and the chaperones involved in their fragmentation. The sizes are drawn to scale. Structures of yeast chaperones are presented, while their mentioned human orthologs are of similar size and structure, such as (A) Sis1/DNAJB1 dimer (Sis1: PDB 6D6X+1CG3). JD: J domain; CTDI, CTDII: carboxy-terminal domains I and II; DD: dimerization domain. (B,C) Ssa1/Hsc70 and Sse1/HSP110 (Ssa1: PDB 2KHO, Sse1: PDB 2QXL). NBD: nucleotide binding domain; SBDα and SBDβ: substrate-binding domain. (D) Hsp104 hexamer (PDB 5KNE). N, M: domains N and M; NBD1, NBD2: nucleotide binding domains 1 and 2. (E) α-syn fibril (PDB 2N0A). (F) Sup35 prion (Sup35C: PDB 1R5B). Sup35 prion structure with two protease-resistant cores is depicted, though in some [PSI+] variants Core 2 is absent and/or additional structures in the M domain are present. For clarity, only four Sup35 C domains and M domains (blue line) are shown; other copies of the M domain are indicated by dotted lines. ChP—a non-structured protease-sensitive region of the Sup35N domain, presumably used for productive binding of Hsp104 via Sis1 and Ssa chaperones. ChN—the region 129–148 of direct but non-productive Hsp104 binding. The diameter of the amyloid core, presumably corresponding to the Sup35 N domain, is about 8–10 nm [53,68]. The unfolded M domain allows the C domain to be widely offset from the fibril core, thus they form a loose halo with a diameter of 60–65 nm [68]. Notably, the M domain can acquire a folded [53] protease-resistant state, at least in a part of the prion population [45,53].