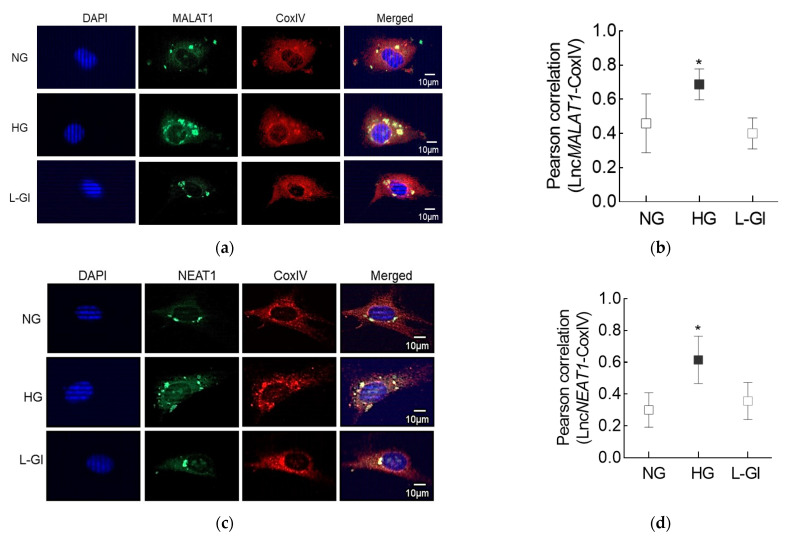
Figure 3.
Effect of high glucose on mitochondrial localization of LncMALAT1 and LncNEAT1. (a,c) Using RNA-FISH technique, cells fixed with 4% paraformaldehyde were incubated with fluorescein-labelled denatured probes. Mitochondrial localization was performed using CoxIV antibody and Texas Red-conjugated secondary antibody. Cells were mounted using DAPI (blue)-containing mounting medium. (b,d) Signals of fluorescein-12-dUTP incorporated probe (green) were visualized to determine the hybridized probes. Pearson’s correlation coefficient of CoxIV with LncMALAT1 or LncNEAT1 was calculated by using the “Region of Interest” in the outer nuclear region from 25 or more cells in each group. Pearson’s correlation coefficient values in the graphs are represented as mean ± SD. NG and HG = Cells in 5 mM or 20 mM d-glucose; L-Gl = cells in 20 mM l-glucose; * p < 0.05 vs. NG.