Figure 1. TAOK2 is an ER-protein kinase.
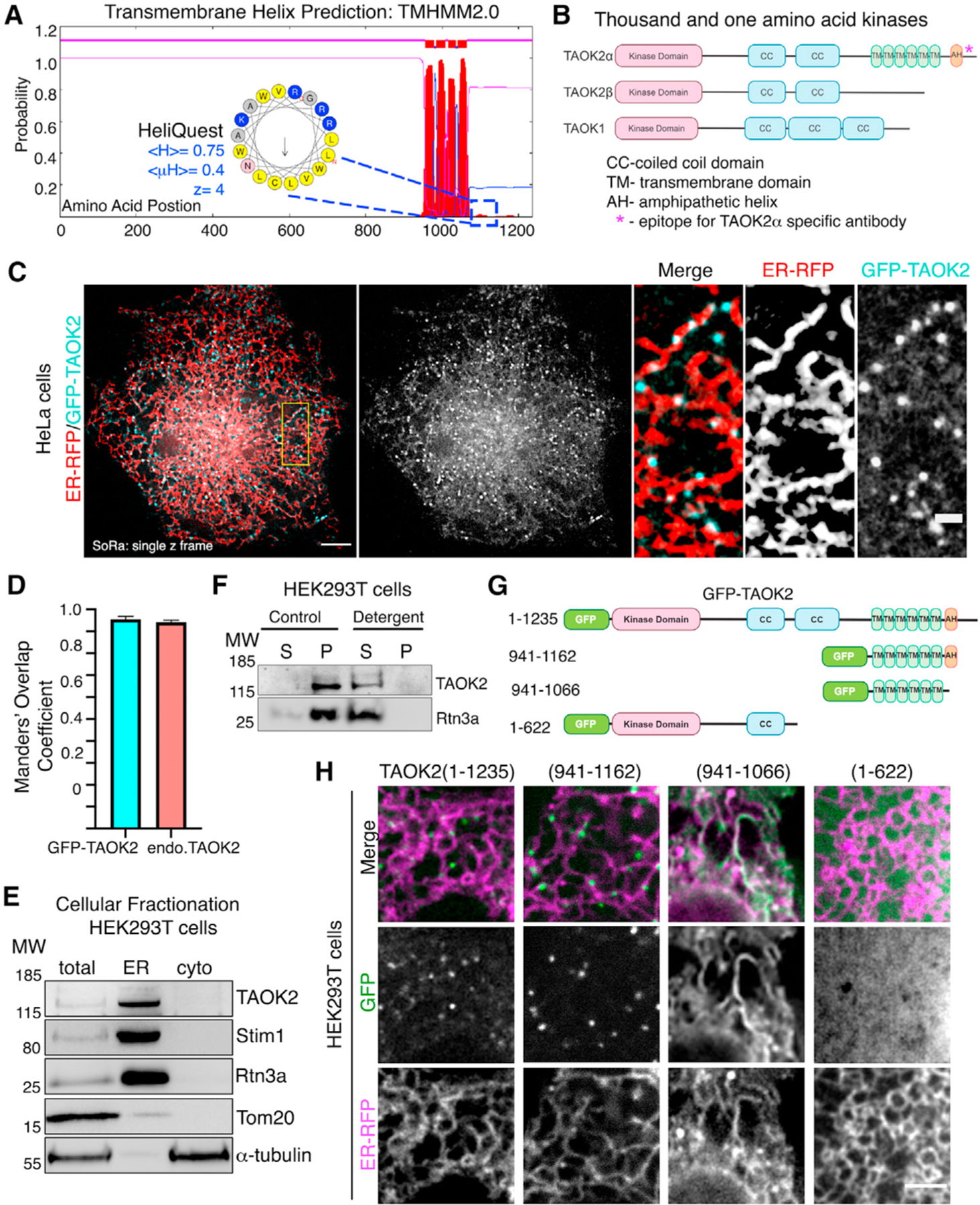
(A) Transmembrane Hidden Markov Model (TMHMM v2.0) prediction plot shows the posterior probabilities (y axis) of inside/outside(magenta)/TM helix (red) along the length of TAOK2 sequence (x axis). Hydrophobic region following the sixth predicted transmembrane domain (TMD) was analyzed through AMPHIPASEEK which predicted residues 1,146–1,162 to be amphipathic. Charged (blue) and hydrophobic (yellow) residues are indicated in the inset. HeliQuest was used to calculate hydrophobicity <H>, hydrophobic moment <μH> and net charge Z.
(B) Schematic representation of TAOK2 isoforms α, β and TAOK1. Coiled coils (CC), six transmembrane domains (TM), and amphipathic helix (AH) predicted in TAOK2 are depicted. The asterisk indicates the epitope used to generate the TAOK2α-specific antibody.
(C) SoRa (super resolution by optical reassignment) images of HeLa cells expressing GFPTAOK2 (cyan) and ER marker ER-mRFP (red). Scale bar: 5 μm. Grayscale images of TAOK2 and ER-RFP are shown separately. Scale bar: 1 μm.
(D) Manders’ overlap coefficient for co-localization of expressed GFP-TAOK2 and ER-mRFP and for endogenous TAOK2 and EGFP-Sec22b are plotted. Values indicate mean ± SEM; n = 10 for each condition.
(E) Western blot (n = 3) of cell homogenate fractionated into ER membrane and cytosol probed with antibodies against TAOK2, ER-membrane proteins Stim1 and Rtn3, mitochondrial protein Tom20, and tubulin.
(F) Western blot (n = 3) of cell homogenate fractionated into membrane pellet (P) and cytosolic supernatant (S) components, in the absence (control) or presence of detergent, probed with antibodies against TAOK2 and ER protein Rtn3a.
(G) GFP-tagged TAOK2 deletion constructs used in (H).
(H) Confocal images of cells transfected with distinct GFP-tagged TAOK2 deletion constructs (green) and ER-mRFP (magenta). Scale bar: 3 μm.
See also Figure S1.
