Figure 6. TAOK2 localizes to the mitotic spindle poles and regulates mitotic ER remodeling.
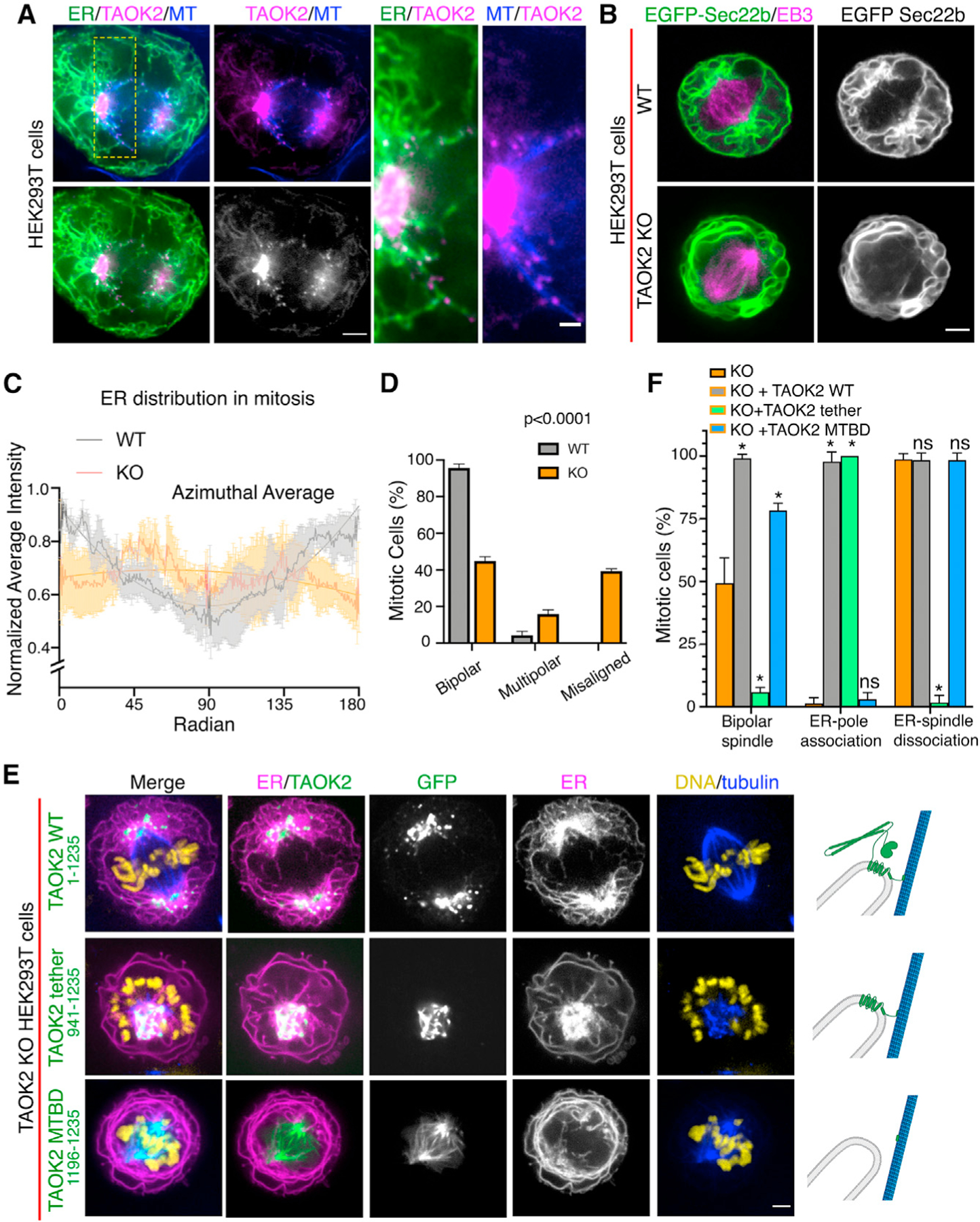
(A) SoRa confocal image of mitotic cell expressing GFP-TAOK2 (magenta) and ER-mRFP (green) and stained with MT dye (blue); scale bar: 2μm. Magnified view of the ER-MT tethering sites; scale bar: 0.5 μm.
(B) WT and TAOK2 KO mitotic cells transfected with EGFP-Sec22b (green) and mCherry-EB3 (magenta). ER morphology in WT and TAOK2 KO cells is shown in grayscale. Scale bar: 5 μm.
(C) Azimuthal average of normalized fluorescence intensity of ER markers (where 0 and 180 correspond to the spindle poles) is plotted for WT (gray) and TAOK2 KO (orange) cells. Values indicate mean ± SEM; n = 3 cells from 3 experiments.
(D) Percent mitotic WT (gray) and TAOK2 KO (orange) cells that exhibit bipolar, multipolar, or misaligned mitotic spindles. Values indicate mean ± SEM; n > 45 mitotic cells from 3 experiments; p < 0.0001 one-way ANOVA.
(E) Mitotic TAOK2 KO cells expressing indicated TAOK2 constructs (green), ER-mRFP (magenta), DNA (yellow), and MT (blue). Localization of TAOK2 constructs and their effect on ER morphology is shown in grayscale. Scale bar: 5 μm. Schematic (right) depicts topology of GFP-TAOK2 deletion constructs.
(F) Percentage of mitotic TAOK2 KO cells expressing the indicated constructs were scored as (1) exhibiting normal bipolar spindle, (2) ER association with spindle poles, and (3) ER dissociation from mitotic spindle. Values indicate mean ± SEM; n = 75 cells from 3 experiments. *p < 0.05.
