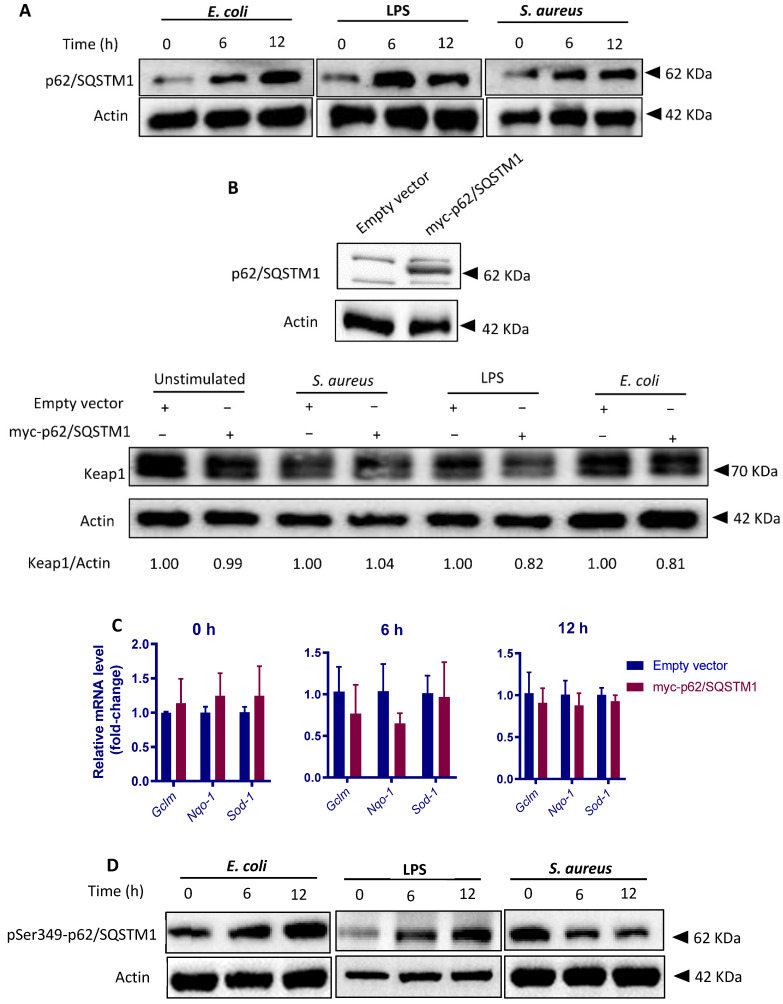
Figure 10.
S. aureus and E. coli differentially modify p62/ SQSTM1 phosphorylation. (A) Evaluation of the p62/SQSTM1 protein levels in pbMECs. Cells were treated with heat-killed E. coli (1 × 107 particles /mL), LPS (10 μg/mL) or S. aureus (1 × 107 particles/mL) for the indicate time. Whole-cell extracts were analysed by immunoblotting with anti-p62/SQSTM1. β-actin is shown as a loading control. The data are representative of 2 independent experiments. (B) Evaluation of the p62/SQSTM1-dependent Keap1 degradation. pbMECs were transfected with either mock (empty vector) or myc-p62/SQSTM1 plasmid. Forty-eight hours post-transfection, the cells were treated heat-killed E. coli (1 × 107 particles /mL), LPS (10 μg/mL) or S. aureus (1 × 107 particles/mL) for a further 6 h. Whole-cell extracts were analysed by immunoblotting with anti-Keap1. β-actin is shown as a loading control. The data are representative of 2 independent experiments. (C) Evaluation of Nrf2 activation in cells with p62/SQSTM1 overexpression in response to S. aureus. Mock-transfected and p62/SQSTM1 plasmid-transfected cells were treated as indicated above. Total mRNA was extracted and RT-qPCR was performed to measure the mRNA levels of Nrf2 target genes Ho-1, Glcm, Nqo-1, and Sod1. Data presented are the mean ± s.d. of three triplicates and are representative of 3 separate experiments. (D) Evaluation of the effect of E. coli, S. aureus and LPS on phosphorylation of p62/SQSTM1 at Ser349 (pSer349-p62/SQSTM1). pbMECs were treated with heat-killed E. coli (1 × 107 particles /mL), LPS (10 μg/mL) or S. aureus (1 × 107 particles/mL) for the indicated time. Whole-cell extracts were analysed by immunoblotting with anti-pSer349-p62/SQSTM1. β-actin is shown as a loading control. The data are representative of 2 independent experiments.