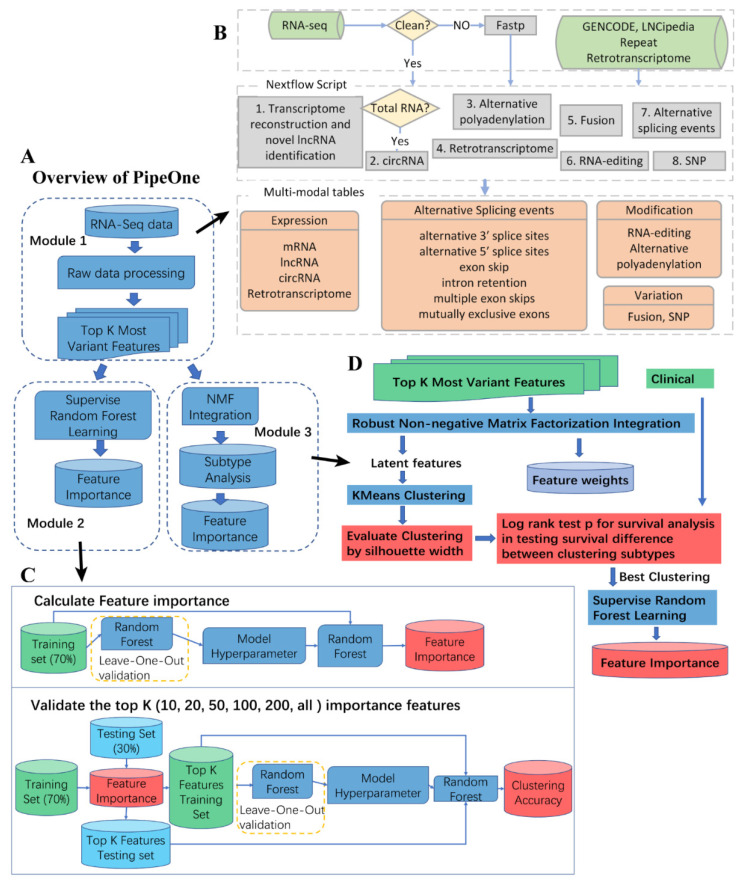
Figure 1.
Overall design of PipeOne. (A) Three modules of PipeOne: data processing and various feature identification (one), feature prioritizing (two), and disease subtyping (three). (B) Details of module one. Raw sequencing reads were quality controlled by FASTP and then went through eight tools to extract information from RNA-seq data, including expression levels of mRNA, lncRNA, circRNA, and retrotransposons, alternative splicing events, alternative polyadenylation, RNA editing, gene fusions, and SNPs. These information was used to construct the feature matrices for machine learning (only the top 1000 most variable features were used for each type of information) in module two and three. (C) Details of module two. First, feature importance was calculated by using random forest on all features from module one. Then the top K (20, 20, 50, 100, 200, all) ranked by feature importance were used to test and validate the importance of those top features. (D) Details of module three. First, a robust NMF integration algorithm was applied to obtain latent features and associated weights for all samples. Then K-Means clustering evaluated by silhouette width was used to cluster samples based on the latent feature matrix. Differential survival analysis by log-rank test was used to assess the clinical relevance of those stable clusters as potential subtypes. Finally, similar to module two, random forest was used to select features contributing to the subtyping results.