FIG. 9.
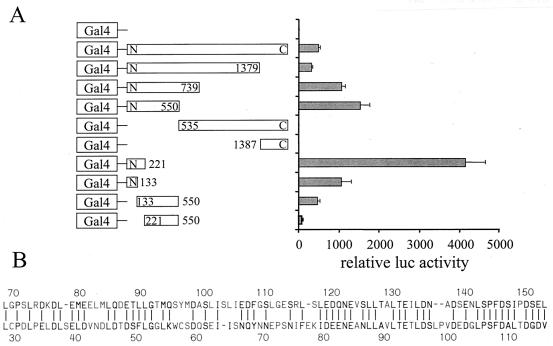
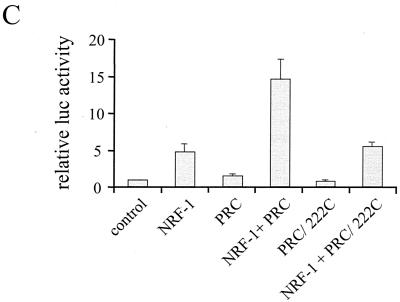
Mapping of the PRC activation domain. (A) BALB/3T3 cells were transfected with a 5xGal4 luciferase reporter plasmid, CMV/β-gal, and either pSG424 control vector (Gal4) or the various PRC/pSG424 fusions, as indicated. Results are expressed as the ratio of the luciferase to β-galactosidase activities and are the means ± standard errors of the mean for at least three independent experiments, each performed in duplicate. (B) Comparison of the putative activation domains in PRC and PGC-1. A Lipman-Pearson protein alignment of aa 69 to 154 of PRC and aa 29 to 117 of PGC-1 is shown. Identical or homologous (by 1 distance unit) amino acids are indicated by a vertical line (|). (C) Comparison of wild-type PRC to the amino-terminal deletion PRC/222C in the coactivation of NRF-1-dependent transcription from a 4xNRF-1/luc reporter plasmid. Results are expressed as in shown in panel A.