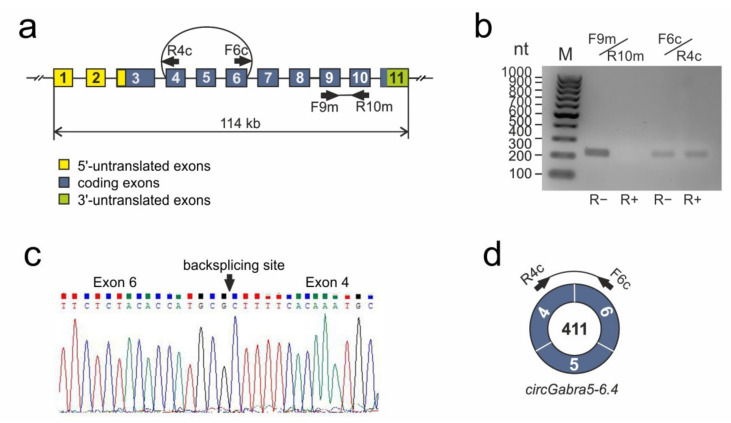
Figure 2.
An analysis of circRNA diversity in tMCAO rat model conditions. (a) The exon-intron structure of the rat Gabra5 gene. Exons were shown as numerated blocs and the separating introns, as straight lines connecting rectangles. Arcs connect the ends of exons involved in the formation of circRNA. Primers (F, forward; R, reverse) are shown as arrows, the direction indicates the 5′-3′-orientation. (b) Electrophoretic separation of the cDNA amplification products of the rat brain in the presence of primers complementary to exon sequences of the Gabra5 gene. On the left, the position of markers bands is indicated by GeneRuler 100 bp DNA Ladder (Thermo Fisher Scientific, Waltham, MA, USA). (c) Sanger sequencing result of circGabra5-6.4 matched to regions of exons that participated in back-splicing. It was visualized using Chromas Lite (Technelysium Pty Ltd, South Brisbane, Australia) software. (d) The structure of circRNA circGabra5-6.4 of the Gabra5 gene, whose formation was confirmed using PCR and Sanger sequencing. The number inside the circle is the length of circRNAs (nucleotides), figures in the circle segments are the numbers of exons inside the circRNAs.