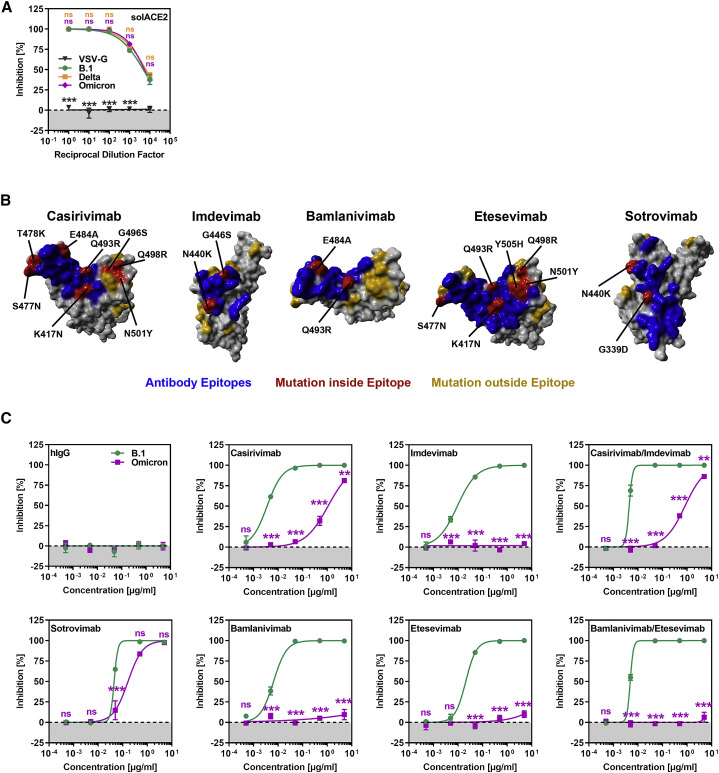
Figure 2.
The Omicron spike evades neutralization by four out of five monoclonal antibodies but is efficiently inhibited by soluble ACE2
(A) Inhibition of S-protein-driven cell entry by soluble ACE2. Particles harboring the indicated S proteins were preincubated (30 min, 37°C) with different dilutions of soluble ACE2 before being inoculated onto Vero cells. S-protein-driven cell entry was analyzed as described in Figure 1F. Presented are the mean data from three biological relocates (each conducted with four technical replicates).
(B) Locations of Omicron-specific mutations in the context of the RBD epitopes targeted by casirivimab, imdevimab, bamlanivimab, etesevimab, and sotrovimab. (RBD, gray; epitope targeted by the antibody, blue; Omicron-specific mutations within the epitope, red; Omicron-specific mutations outside the epitope, orange).
(C) Omicron spike is resistant against four out of five monoclonal antibodies used for treatment of COVID-19 patients. Particles harboring the indicated S proteins were preincubated (30 min, 37°C) with different concentrations of recombinant monoclonal antibody before being inoculated onto Vero cells. S-protein-driven cell entry was analyzed as described in Figure 1F. Presented are the mean data from three biological replicates (each conducted with four technical replicates).
(A and C): statistical significance of differences between individual groups was assessed by two-way analysis of variance with Dunnett’s (A) or Sidak’s (C) post hoc tests: p > 0.05, not significant (ns); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.