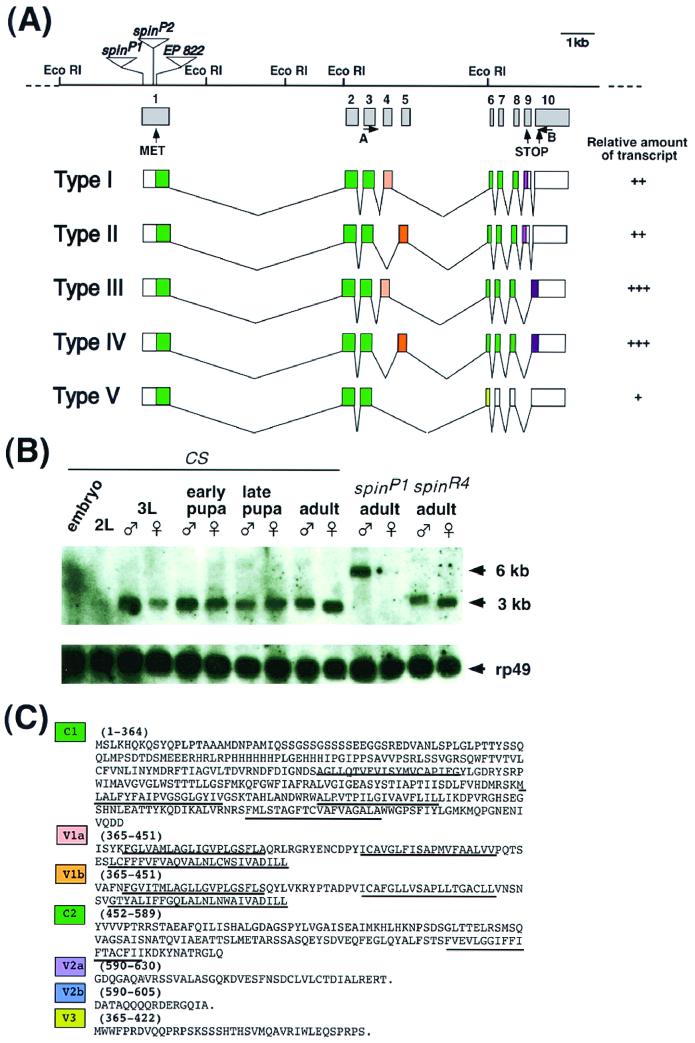
FIG. 6.
Molecular characterization of the spin gene and its products. (A) Genomic structure of the spin gene and the structures of five transcripts produced by alternative splicing. The restriction sites are shown for the EcoRI restriction enzyme. The P-element insertion site in the spinP1 mutant is located 8 bp downstream of the transcription initiation site and is indicated by a triangle. The enhancer trap vectors in the spinP2 and EP822 genes are inserted in the middle of exon 1. The relative amount of each type of transcript is represented as follows: +++, abundant; ++; modest; or +, rare. No obvious differences were found during development between the sexes or between wild-type and spinP1 mutant flies. The primers used for RT-PCR were primers A and B. (B) Developmental Northern blots probed by spin cDNA. Twenty microgram samples of poly(A)+ RNAs extracted from embryos, second-instar larvae, third-instar larvae, early pupae, late pupae, adults of the wild type, adults with the spinP1 mutation, and adults with the revertant spinR4 mutation were loaded onto the gel. The blots were then probed with the spin type I cDNA as well as the ribosomal protein gene rp49 cDNA in order to assess the amount of RNA loaded in each lane. (C) Amino acid sequences of each of the domains of Spin. C1 and C2 represent common domains. The V1, V2, and V3 domains are variable (see panel A). The predicted transmembrane domains are underlined.