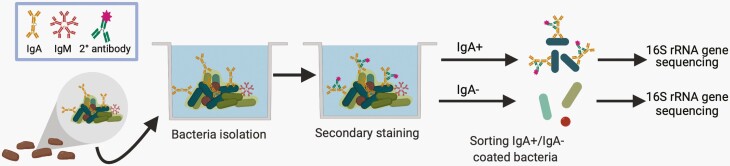
Figure 1.
Schematic of mFLOW-Seq for identifying fecal microbiota coated by mucosal IgA. Bacteria isolated from the feces are stained with a fluorescently labeled anti-IgA secondary antibody. Then fluorescence-activated cell sorting (FACS) is used to separate the IgA+ and IgA− bacteria. The IgA+ and IgA− populations are then analyzed by 16S rRNA gene sequencing to determine which microbes are enriched in the IgA+ fraction as an indicator of those microbes selectively bound by secretory IgA.