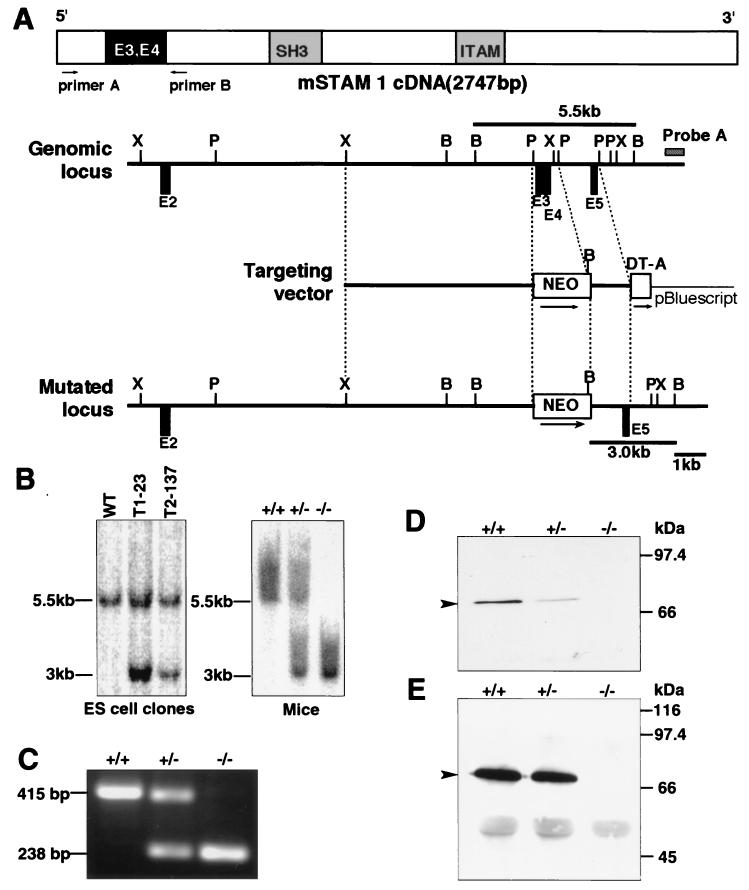
FIG. 1.
Generation of STAM1-deficient mice. (A) Schematic representation of the mouse STAM1 (mSTAM1) cDNA, stam1 genomic locus, targeting vector, and stam1 mutated locus. The positions of stam1 exons are shown as boxes. The targeting vector was designed to replace exon 3 (E3) and E4, encoding amino acids 42 to 99 of STAM1. The fragments expected to be generated by BamHI digestion are 5.5 and 3.0 kb for the wild-type and the mutated alleles, respectively. B, BamHI; P, PstI; X, XbaI. (B) Southern blot analysis of the stam1 mutation in ES cell clones and mice. Lines indicate the positions of the DNA fragments corresponding to the wild-type (5.5 kb) and mutated (3.0 kb) alleles. (C) RT-PCR analysis of total RNA from purified splenocytes of STAM1+/+, STAM1+/−, and STAM1−/− mice. The primers used are primers A and B, shown in panel A. (D) Western blot analysis for STAM1. Neocortex lysates (20 μg) from STAM1+/+, STAM1+/−, and STAM1−/− mice were separated by SDS-PAGE and blotted with anti-STAM1 antibody. The position of STAM1 is marked by an arrowhead. (E) Immunoprecitipation analysis for STAM1. Lysates of activated T cells from STAM1+/+, STAM1+/−, and STAM1−/− mice were immunoprecipitated and then immunoblotted with anti-STAM1 antibody. The position of STAM1 is marked by an arrowhead.