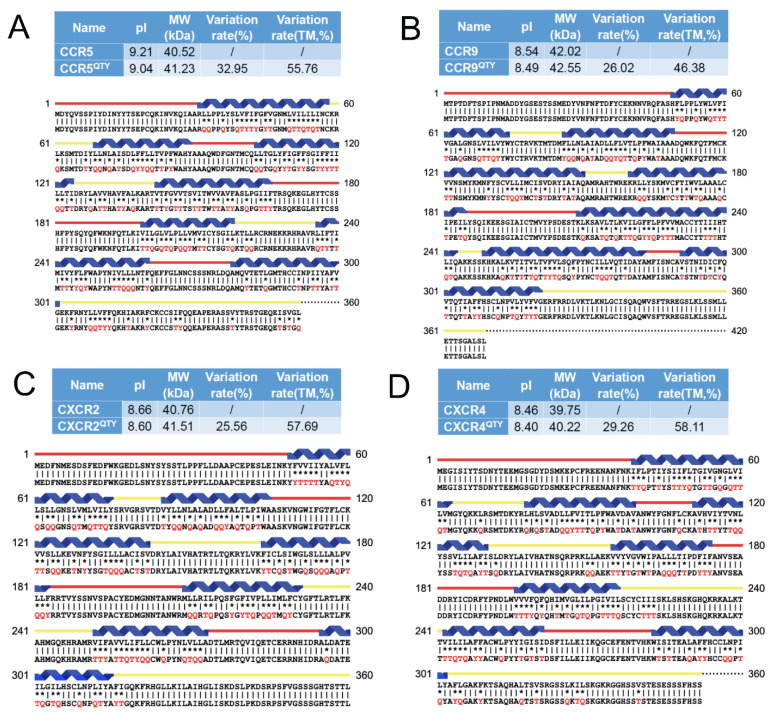
Figure 1.
Alignment of known X-ray crystal structures of native chemokine receptors CCR5, CCR9, CXCR2 and CXCR4 with AlphaFold2 predicted native and their predicted water-soluble QTY variants. The Q, T, and Y amino acid substitutions are in red. The alpha-helical segments (blue) are shown above the protein sequences, the external (red) and internal (yellow) loops of the receptors are indicated. The symbols | and * indicate the similar and different amino acids, respectively. Characteristics of natural and QTY variants with pI, molecular weight, total variation rate and membrane variation rate, and the alignment: (A) CCR5 and CCR5QTY, (B) CCR9 and CCR9QTY, (C) CXCR2 and CXCR2QTY, and (D) CXCR4 and CXCR4QTY. Since the internal regions ICL1, ICL2, ICL3 and the C-terminus do not interact with the ligands, additional residues in these regions are sometimes QTY modified.