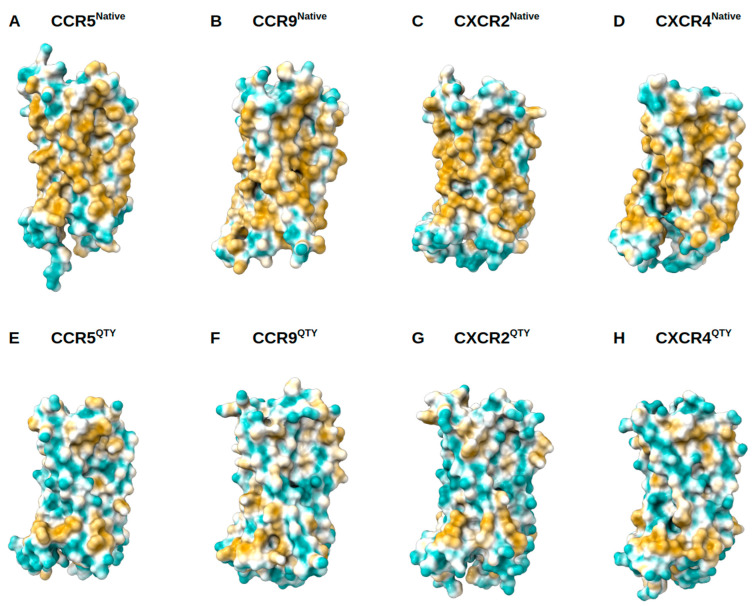
Figure 5.
Surface hydrophobic patch of X-ray determined structures of native chemokine receptors and AlphaFold2 predicted water-soluble QTY variants. The native GPCR receptors mostly expose hydrophobic residues leucine (L), isoleucine (I), valine (V) and phenylalanine (F) facing outside to the hydrophobic lipid bilayer in cell membrane. After replacing the L, I, V, F with polar amino acids, glutamine (Q), threonine (T), and tyrosine (Y), the surfaces are much less hydrophobic. The large surface hydrophobic patch (yellow color) of the native receptors determined by X-ray crystal structures: (A) CCR5, (B) CCR9, (C) CXCR2 and (D) CXCR4. The hydrophobic patch is significantly reduced on the transmembrane domains for the AlphaFold2 predicted water-soluble QTY variants: (E) CCR5QTY, (F) CCR9QTY, (G) CXCR2QTY, (H) CXCR4QTY. These QTY variants become water-soluble without any detergent. The N- and C-termini are removed for clarity of directly comparisons.