Table 1.
Pharmacogenetics of selected drug categories with effects on the cardiovascular system.
| Agents Acting on the Renin–Angiotensin System | ||
|---|---|---|
| ACE Inhibitors | ||
| Drug | Properties | Pharmacogenetics |
|
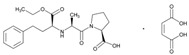
Name: ENALAPRIL IUPAC Name: L-proline, 1-[N-[1-(ethoxycarbonyl)-3-phenylpropyl]-L-alanyl]-, (S)-, (Z)-2-butenedioate (1:1); 1-[N-[(S)-1-carboxy-3-phenylpropyl]-L-alanyl]-L-proline 1′-ethyl ester, maleate (1:1) Molecular Formula: C20H28N2O5 C4H4O4 Molecular Weight: 492.52 Mechanism: Competitive inhibitor of angiotensin-converting enzyme (ACE). Prevents conversion of angiotensin I to angiotensin II, a potent vasoconstrictor. Results in lower levels of angiotensin II, which causes an increase in plasma renin activity and a reduction in aldosterone secretion. Effect: Treatment of hypertension, symptomatic heart failure, or asymptomatic left ventricular dysfunction. |
Mechanistic genes:ACE1, ADRB2, AGTR1, AGT, BDKRB2, NOS3 Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:ACE1 Transporter genes:SLC22A8 Pleiotropic genes:IL6 |
|
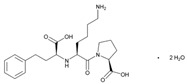
Name: LISINOPRIL IUPAC Name: L-proline, 1-[N2-(1-carboxy-3-phenylpropyl)-L-lysyl]-, dihydrate, (S)-; 1-[N2-[(S)-1-carboxy-3-phenylpropyl]-L-lysyl]-L-proline dihydrate Molecular Formula: C21H31N3O5 2H2O Molecular Weight: 441.52 Mechanism: Competitive inhibitor of angiotensin-converting enzyme (ACE). Prevents conversion of angiotensin I to angiotensin II, a potent vasoconstrictor. Effect: Treatment of hypertension, either alone or in combination with other antihypertensive agents. Adjunctive therapy in treatment of heart failure. Treatment of acute MI within 24 h in hemodynamically stable patients to improve survival. |
Mechanistic genes:ACE1, ADD1, AGTR1, AGT Metabolic genes Substrate:ACE1, ADD1, AGTR1, AGT Inhibitor:ACE1, ACE2 |
|
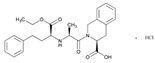
Name: QUINAPRIL IUPAC Name: 3-Isoquinolinecarboxylic acid, 2-[2-[[1-(ethoxycarbonyl)-3-phenylpropyl]amino]-1-oxopropyl]-1,2,3,4-tetrahydro-, monohydrochloride, [3S-[2[R*(R*)],3R*]]; (S)-2-[(S)-N-[(S)-1-carboxy-3-phenylpropyl]alanyl]-1,2,3,4-tetrahydro-3-isoquinolinecarboxylic acid, 1-ethyl ester, monohydrochloride Molecular Formula: C25H30N2O5 HCl Molecular Weight: 474.98 Mechanism: Treatment of hypertension. Reduction in cardiovascular mortality or non-fatal myocardial infarction in stable coronary artery disease. Effect: Hypertension. Heart failure. |
Mechanistic genes: ACE1, AGT, AGTR1, BDKBR2, NR1I21 TGFB1 Metabolic genes Substrate: CYP11B2 Inhibitor: ACE1 |
|
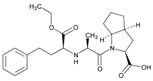
Name: RAMIPRIL IUPAC Name: [1,1′-Biphenyl]-2-carboxylic acid, 4′-[(1,4′-dimethyl-2′-propyl[2,6′-bi-1H-benzimidazol]-1′-yl)methyl]-; 4′-[[4-methyl-6-(1-methyl-2-benzimidazolyl)-2-propyl-1-benzimidazolyl]methyl]-2-biphenylcarboxylic acid Molecular Formula: C23H32N2O5 Molecular Weight: 514.62 Mechanism: A non-peptide AT1 angiotensin II receptor antagonist. This binding prevents angiotensin II from binding to receptor thereby blocking vasoconstriction and aldosterone-secreting effects of angiotensin II. Effect: Treatment of hypertension, alone or in combination with other antihypertensive agents. |
Mechanistic genes: ACE1, AGT, AGTR1, BDKRB2, ERAP1, PPARG Metabolic genes Substrate: CYP2C9, CYP11B2, UGT1A1 Inhibitor: ABCB1, ABCG2, CYP2C9, CYP2C19 |
| Angiotensin II Antagonists | ||
|
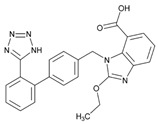
Name: CANDESARTAN IUPAC Name: 1H-benzimidazole-7-carboxylic acid, 2-ethoxy-1-[[2′-(1H-tetrazol-5-yl)[1,1′-biphenyl]-4-yl]methyl]-; 2-ethoxy-1-[p-(o-1H-tetrazol-5-ylphenyl)benzyl]-7-benzimidazolecarboxylic acid Molecular Formula: C24H20N6O3 Molecular Weight: 440.45 Mechanism: Candesartan is an angiotensin receptor antagonist, blocking vasoconstriction and the aldosterone-secreting effects (reabsorption of sodium and water) of angiotensin II. Effect: Essential hypertension. Heart failure. |
Metabolic genes Substrate: CYP1A1, CYP2C9, CYP11B2, UGT1A3, UGT1A5, UGT2B7 Inhibitor: ABCG2, CYP2C8, CYP2C9 Transporter genes:ABCB1, ABCG2 |
|
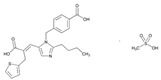
Name: EPROSARTAN IUPAC Name: 2-Thiophenepropanoic acid, α-[[2-butyl-1-[(4-carboxyphenyl)methyl]-1H-imidazol-5-yl]methylene]-, €-, monomethanesulfonat€(E)-2-butyl-1-(p-carboxybenzyl)-α-2-thenylimidazole-5-acrylic acid, monomethanesulfonate Molecular Formula: C23H24N2O4S CH4O3S Molecular Weight: 520.62 Mechanism: A non-biphenyl, non-tetrazole angiotensin II receptor (AT1) antagonist. Blocks the vasoconstrictor and aldosterone-secreting effects of angiotensin II by selectively blocking the binding of angiotensin II to the AT1 receptor in many tissues, such as vascular smooth muscle and adrenal gland. Does not bind to or block other hormone receptors or ion channels known to be important in cardiovascular regulation. Effect: Used alone or in combination with other classes of antihypertensive agents in the management of hypertension. |
Mechanistic genes: ACE1, AGTR1 Metabolic genes Inhibitor: CYP2C9 Inducer: ABCC2 Transporter genes: ABCB1, ABCG2 |
|
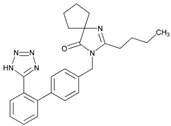
Name: IRBESARTAN IUPAC Name: 1,3-Diazaspiro[4.4]non-1-en-4-one, 2-butyl-3-[[2′-(1H-tetrazol-5-yl)[1,1′-biphenyl]-4-yl]methyl]-; 2-butyl-3-[p-(o-1H-tetrazol-5-ylphenyl)benzyl]-1,3-diazaspiro[4.4]non-1-en-4-one Molecular Formula: C25H28N6O Molecular Weight: 428.53 Mechanism: Irbesartan binds to AT1 angiotensin II receptor. This binding prevents angiotensin II from binding to receptor, thereby blocking the vasoconstriction and aldosterone-secreting effects of angiotensin II. Effect: Treatment of hypertension alone or in combination with other antihypertensives. Treatment of diabetic nephropathy in type 2 diabetes mellitus (non-insulin-dependent, NIDDM) and hypertension. |
Mechanistic genes: ADRA1A, AGTR1, APOB, BDKRB2, ERAP1, EDN1, NPPA, AGT, APOE, LDLR, NOS3, TGFB1 Metabolic genes Substrate: CYP2C9, CYP3A4, CYP3A5, CYP11B2 Inhibitor: CYP1A2, CYP2C8, CYP2C9, CYP2D6, CYP3A4, CYP3A5 Transporter genes: ABCB1, ABCG2 |
|
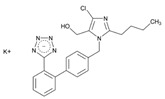
Name: LOSARTAN IUPAC Name: 1H-Imidazole-5-methanol, 2-butyl-4-chloro-1-[[2′-(1H-tetrazol-5-yl)[1,1′-biphenyl]-4-yl]methyl]-, monopotassium salt; 2-butyl-4-chloro-1-[p-(o-1H-tetrazol-5-ylphenyl)benzyl]imidazole-5-methanol, monopotassium salt Molecular Formula: C22H22ClKN6O Molecular Weight: 461.00 Mechanism: As a selective and competitive non-peptide angiotensin II receptor antagonist, losartan blocks vasoconstrictor and aldosterone-secreting effects of angiotensin II. Losartan increases urinary flow rate and in addition to being natriuretic and kaliuretic, increases excretion of chloride, magnesium, uric acid, calcium, and phosphate. Effect: Treatment of hypertension. Treatment of diabetic nephropathy in type 2 diabetes mellitus (non-insulin-dependent) and history of hypertension. Stroke risk reduction in hypertension and left ventricular hypertrophy. |
Mechanistic genes:ACE1, ADD1, AGT, AGTR1, AGTR2, BDKRB2, EDN1, FOS, MMP2, NOS3, PDGFRB, REN, TGFB1 Metabolic genes Substrate:CYP1A2, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, UGT1A1, UGT1A3, UGT1A10, UGT2B7, UGT2B17 Inhibitor:CYP1A2, CYP2C8, CYP2C9, CYP2C19, CYP3A4, CYP3A5, CYP11B2 Transporter genes:ABCB1, ABCG2 Pleiotropic genes:TNF |
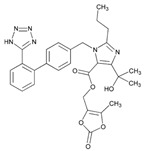
|
Name: OLMESARTAN IUPAC Name: 1H-Imidazole-5-carboxylic acid, 4-(1-hydroxy-1-methylethyl)-2-propyl-1-[[2′-(1H-tetrazol-5-yl) [1,1′-biphenyl]-4-yl]methyl]-, (5-methyl-2-oxo-1,3-dioxol-4-yl) methyl ester Molecular Formula: C29H30N6O6 Molecular Weight: 558.59 Mechanism: Blocks vasoconstrictor and aldosterone-secreting effects of angiotensin II. Interacts reversibly at AT1 and AT2 receptors and has slow dissociation kinetics (has greater affinity for AT1 receptor). Olmesartan increases urinary flow rate and, besides being natriuretic and kaliuretic, increases excretion of chloride, magnesium, uric acid, calcium, and phosphate. Effect: Hypertension. |
Mechanistic genes:ACE2, AGTR1, EDN1, TGFB1 Metabolic genes Substrate:CMBL, CYP2C9 Inducer:ABCC2 Transporter genes:ABCB1, ABCC2, ABCG2, SLC22A8, SLCO1A2, SLCO1B1 Pleiotropic genes:APOE |
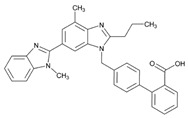
|
Name: TELMISARTAN IUPAC Name: [1,1′-Biphenyl]-2-carboxylic acid, 4′-[(1,4′-dimethyl-2′-propyl[2,6′-bi-1H-benzimidazol]-1′-yl)methyl]-; 4′-[[4-methyl-6-(1-methyl-2-benzimidazolyl)-2-propyl-1-benzimidazolyl]methyl]-2-biphenylcarboxylic acid Molecular Formula: C33H30N4O2 Molecular Weight: 514.62 Mechanism: A non-peptide AT1 angiotensin II receptor antagonist. This binding prevents angiotensin II from binding to receptor thereby blocking vasoconstriction and aldosterone-secreting effects of angiotensin II. Effect: Treatment of hypertension, alone or in combination with other antihypertensive agents. |
Mechanistic genes:ACE1, AGT, AGTR1, BDKRB2, ERAP1, PPARG Metabolic genes Substrate:CYP2C9, CYP11B2, UGT1A1 Inhibitor:ABCB1, ABCG2, CYP2C9, CYP2C19 |
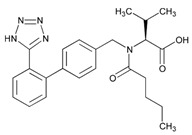
|
Name: VALSARTAN IUPAC Name: L-valine, N-(1-oxopentyl)-N-[[2′-(1H-tetrazol-5-yl)[1,1′-biphenyl]-4-yl]methyl]-; N-[p-(o-1H-tetrazol-5-ylphenyl)benzyl]-N-valeryl-L-valine Molecular Formula: C24H29N5O3 Molecular Weight: 435.52 Mechanism: Displaces angiotensin II from AT1 receptor and produces its blood pressure-lowering effects by antagonizing AT1-induced vasoconstriction, aldosterone release, catecholamine release, arginine vasopressin release, water intake, and hypertrophic responses. Effect: Treatment of essential hypertension (alone or in combination with other antihypertensive agents). Reduction in cardiovascular mortality in left ventricular dysfunction postmyocardial infarction. Treatment of heart failure. |
Mechanistic genes:ACE1, AGT, AGTR1, AGT2R1, BDKRB2, ERAP1, GNB3, STAT3, TGFB1 Metabolic genes Substrate:CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, CYP11B2 Inhibitor:CYP2C9 Transporter genes:ABCC2, SLCO1B1, SLCO1B3 |
| Other Agents acting on the Renin–Angiotensin system | ||
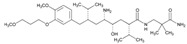
|
Name: ALISKIREN IUPAC Name: Benzeneoctanamide, δ-amino-N-(3-amino-2,2-dimethyl-3-oxopropyl)-γ-hydroxy-4-methoxy-3-(3-methoxypropoxy)-α,ζ-bis(1-methylethyl)-, (αS, γS, δS, ζS)-; (2) (2S,4S,5S,7S)-5-amino-N-(2-carbamoyl-2-methylpropyl)-4-hydroxy-2-isopropyl-7-[4-methoxy-3-(3-methoxypropoxy)benzyl]-8-methylnonamide Molecular Formula: C30H53N3O6 Molecular Weight: 551.76 Mechanism: Blocks conversion of angiotensinogen to angiotensin I. Effect: Treatment of hypertension. |
Mechanistic genes:REN Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:CYP3A4, CYP3A5, REN Transporter genes:ABCB1 Mechanistic genes:ACE1, ACE2, ADD1, ADRB2, AGT, AGTR1, MTHFR, MTR |
| Antihypertensives | ||
| Drug | Properties | Pharmacogenetics |
|
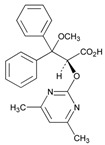
Name: AMBRISENTAN IUPAC Name: (+)-(2S)-2-[(4,6-dimethylpyrimidin-2-yl)oxy]-3-methoxy-3,3-diphenylpropanoic acid Molecular Formula: C22H22N2O4 Molecular Weight: 378.42 Mechanism: Blocks endothelin receptor ETA and ETB on vascular endothelium and smooth muscle. Effect: Treatment of pulmonary artery hypertension. |
Mechanistic genes:EDN1, EDNRA, NOS3 Metabolic genes Substrate:CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTs, UGT1A3, UGT1A9, UGT2B7 Transporter genes:ABCB1, SLCO1A2 Pleiotropic genes:IL1B, IL6 |
|
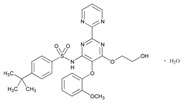
Name: BOSENTAN IUPAC Name: Benzenesulfonamide, 4-(1,1-dimethylethyl)-N-[6-(2-hydroxyethoxy)-5-(2-methoxyphenoxy)[2,2′-bipyrimidin]-4-yl]-, monohydrate; (2) p-tert-butyl-N-[6-(2-hydroxyethoxy)-5-(o-methoxyphenoxy)-2-(2-pyrimidinyl)-4-pyrimidinyl]benzenesulfonamide monohydrate Molecular Formula: C27H29N5O6S.H2O Molecular Weight: 569.63 Mechanism: Acts as a competitive antagonist and blocks endothelin receptors on vascular endothelium and smooth muscle. Stimulation of endothelin receptors is associated with vasoconstriction and proliferation. Although bosentan blocks both ETA and ETB receptors, the affinity is slightly higher for ETA. Effect: Adjunctive therapy for the treatment of pulmonary arterial hypertension (WHO group I), in patients with WHO class III or IV symptoms. |
Metabolic genes Substrate:ACVRL1, BMPR2, EDNRA, EDNRB, TGFBR1 Inhibitor:CYP2B6, CYP2C9, CYP3A4, CYP3A5 Inducer:CYP2C9, CYP2C19, CYP3A4, CYP3A5 Transporter genes:ABCB1, ABCB11, SLCO1B1, SLCO1B3 Pleiotropic genes:TNF |
|
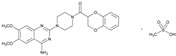
Name: DOXAZOSIN IUPAC Name: Piperazine, 1-(4-amino-6,7-dimethoxy-2-quinazolinyl)-4-[(2,3-dihydro-1,4-benzodioxin-2-yl)carbonyl]-, monomethanesulfonate; 1-(4-amino-6,7-dimethoxy-2-quinazolinyl)-4-(1,4-benzodioxan-2-ylcarbonyl)piperazine monomethanesulfonate Molecular Formula: C23H25N5O5.CH4O3S Molecular Weight: 547.58 Mechanism: Doxazosin is a quinazoline-derivative postsynaptic α1-adrenergic blocking agent. It reduces peripheral vascular resistance and blood pressure as a result of its vasodilating effects. The drug produces both arterial and venous dilation. Effects of doxazosin on the cardiovascular system are mediated by the drug’s activity at α1-receptor sites on vascular smooth muscle. Because of the prevalence of α receptors on the prostate capsule, prostate adenoma, and the bladder trigone and the relative absence of these receptors on the bladder body, α-blockers decrease urinary outflow resistance in men. Doxazosin may improve to a limited extent the serum lipid profile and can reduce blood glucose and serum insulin concentrations. The drug does not appear to affect plasma renin activity appreciably. Effect: Treatment of hypertension alone or in conjunction with diuretics, ACE inhibitors, β-blockers, or calcium antagonists. Treatment of urinary outflow obstruction and/or obstructive and irritative symptoms associated with BPH; can be used in combination with finasteride. |
Mechanistic genes:ACE1, ADD1, ADRA1A Metabolic genes Substrate:CYP2C19, CYP2D6, CYP3A4, CYP3A5 Transporter genes:ABCB1 |
|
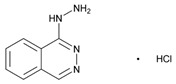
Name: HYDRALAZINE IUPAC Name: Phthalazine, 1-hydrazino-, monohydrochloride; 1-hydrazinophthalazine monohydrochloride Molecular Formula: C8H8N4 HCl Molecular Weight: 196.64 Mechanism: Direct vasodilation of arterioles (with little effect on veins) with decreased systemic resistance. Effect: Management of moderate-to-severe hypertension, congestive heart failure, hypertension secondary to pre-eclampsia/eclampsia. Treatment of primary pulmonary hypertension. |
Mechanistic genes:AGPAT2, AGT, AKR1C4, CHRNA1, COL1A1, ESR1, GSTP1, HBB, HFE, HIF1A, MAOA, MGMT, NR3C1, PDGFRB Metabolic genes Substrate:NAT2 Inhibitor:CEL, CYP3A4, CYP3A5 Transporter genes:SLC6A2, SLC12A3, SLC22A16 Pleiotropic genes:APC, HLA-A, HLA-B, IL6, IL10, TNF, TP53 |
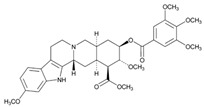
|
Name: RESERPINE IUPAC Name: Yohimban-16-carboxylic acid, 11,17-dimethoxy-18-[(3,4,5-trimethoxybenzoyl)oxy]-, methyl ester, (3β,16β,17α,18β,20α)-; methyl 18β-hydroxy-11,17α-dimethoxy-3β,20α-yohimban-16β-carboxylate 3,4,5-trimethoxybenzoate (ester) Molecular Formula: C33H40N2O9 Molecular Weight: 608.68 Mechanism: Reduces blood pressure via depletion of sympathetic biogenic amines (norepinephrine and dopamine). This also commonly results in sedative effects. Effect: Management of mild-to-moderate hypertension. Treatment of agitated psychotic states (schizophrenia). |
Mechanistic genes:COMT, ERBB2, LDLR, MAOA, MAOB, NR1I2 Metabolic genes Substrate:CYP1A1, CYP3A4, CYP3A5, CYP7A1, UGT1A1 Inhibitor:ABCB1, ABCG2 Inducer:ABCB1 Transporter genes:ABCB11, SLC18A2 |
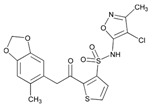
|
Name: SITAXENTAN IUPAC Name: N-(4-chloro-3-methyl-5-isoxazolyl)-2-[[4,5-(methylenedioxy)-o-toly]acetyl]-3-thiophenesulfonamide Molecular Formula: C18H15ClN2O6S2 Molecular Weight: 454.90 Mechanism: A selective antagonist of A subtype of endothelin-1 receptors (ETA) located in pulmonary smooth muscle. Stimulation of these receptors by endogenous endothelin-1 causes vasoconstriction. Sitaxsentan exhibits 6500-fold greater selectivity for ETA over the ETB subtype; the latter predominates on vascular endothelial cells. Thus, preferential antagonism of ETA reduces vasoconstriction, without compromising vasodilatory/antiproliferative actions mediated through endothelin-1 binding to ETB subtype. Effect: Primary pulmonary arterial hypertension or pulmonary hypertension secondary to connective tissue disease. |
Mechanistic genes:EDNRA, EDNRB Metabolic genes Substrate:CYP2C9, CYP3A4, CYP3A5 Inhibitor:CYP2C9, CYP2C19, CYP3A4, CYP3A5 Transporter genes:ABCB1 |
| Beta Blocking Agents | ||
| Drug | Properties | Pharmacogenetics |
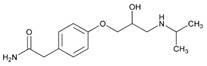
|
Name: ATENOLOL IUPAC Name: Benzeneacetamide, 4-[2-hydroxy-3-[(1-methylethyl)amino]propoxy]-; 2-[p-[2-hydroxy-3-(isopropylamino)propoxy]phenyl]acetamide Molecular Formula: C14H22N2O3 Molecular Weight: 266.34 Mechanism: Competitively blocks response to β-adrenergic stimulation, selectively blocks β1 receptors with little or no effect on β2 receptors except at high doses. Effect: Treatment of hypertension. Management of angina pectoris, postmyocardial infarction. |
Mechanistic genes:ACE1, ACE2, ADRB1, ADRB2, AGT, APOB, BDKRB2, EDN1, ERAP1, GNAS, GNB3, GRK5, LDLR Metabolic genes Substrate:CYP2C9 |
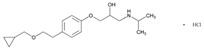
|
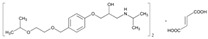
Name: BETAXOLOL IUPAC Name: 2-Propanol, 1-[4-[2-(cyclopropylmethoxy)ethyl]phenoxy]-3-[(1-methylethyl)amino]-, hydrochloride, (±)-; (2)(±)-1-[p-[2-(cyclopropylmethoxy)ethyl]phenoxy]-3-(isopropylamino)-2-propanol hydrochloride Molecular Formula: C18H29NO3 HCl Molecular Weight: 343.89 Mechanism: Competitively blocks β1 receptors, with little or no effect on β2 receptors (bronchial and vascular smooth muscle; only at high doses). No intrinsic sympathomimetic activity and little or no membrane-stabilizing effect (local anesthetic) on the heart. Reduces blood pressure by decreasing cardiac output, decreasing sympathetic outflow from the CNS, and/or suppressing renin release. Reduces intraocular pressure by reducing the production of aqueous humor (may block endogenous catecholamine-stimulated increases in cyclic adenosine monophosphate concentrations within the ciliary processes and subsequent formation of aqueous humor). Effect: Reduction in elevated intraocular pressure in chronic open-angle glaucoma and ocular hypertension (has been used effectively in glaucoma following laser trabeculoplasty). Reduction in systemic hypertension. Initial management of hypertension in heart failure, postmyocardial infarction, high coronary disease risk, and/or diabetes mellitus. |
Mechanistic genes:ADRB1, ADRB2, AGT, BDKRB2, GNAS Metabolic genes Substrate:CYP1A2, CYP2D6 Inhibitor:CYP2D6 |
|
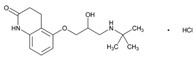
Name: BISOPROLOL IUPAC Name: 2-Propanol, 1-[4-[[2-(1-methylethoxy)ethoxy]methyl]phenoxy]-3-[(1-methylethyl)amino]-, (±)-, I-2-butenedioate (2:1); (2)(±)-1-[[α-(2-Isopropoxyethoxy)-p-tolyl]oxy]-3-(isopropylamino)-2-propanol fumarate (2:1) Molecular Formula: (C18H31NO4)2 C4H4O4 Molecular Weight: 766.96 Mechanism: Selective inhibitor of β1-adrenergic receptors (competitively blocks β1 receptors in myocardium), with little or no effect on β2 receptors at doses ≤20 mg (at high doses may block β2-adrenergic receptors within the bronchial and vascular smooth muscle). Decreases resting and exercise-stimulated heart rate and cardiac output, decreases isoproterenol-induced tachycardia, prolongs sinus node recovery time, refractory period of the AV node, and AV nodal conduction (with rapid atrial stimulation). No intrinsic sympathomimetic activity or membrane-stabilizing effect on the heart. Reduces blood pressure by decreasing cardiac output, decreasing sympathetic outflow from the CNS, and/or suppressing renin release. Effect: Treatment of hypertension, alone or in combination with other agents. Management of mild to moderately severe heart failure of ischemic or cardiomyopathic origin in conjunction with other agents (do not use in patients with acutely decompensated heart failure requiring I.V. inotropic therapy, those with substantial fluid retention requiring intensive diuresis, and those who require hospitalization for heart failure). |
Mechanistic genes:ACE1, ADRB1, AGT, BDKRB2, GNAS Metabolic genes Substrate:CYP2D6, CYP3A4, CYP3A5 |
|
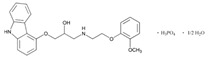
Name: CARTEOLOL IUPAC Name: 2(1H)-quinolinone, 5-[3-[(1,1-dimethylethyl)amino]-2-hydroxypropoxy]-3,4-dihydro-, monohydrochloride; 5-[3-(tert-Butylamino)-2-hydroxypropoxy]-3,4-dihydrocarbostyril monohydrochloride Molecular Formula: C16H24N2O3 HCl Molecular Weight: 328.83 Mechanism: Blocks both β1 and β2 receptors and has mild intrinsic sympathomimetic activity. Reduces intraocular pressure by decreasing aqueous humor production. Has negative inotropic and chronotropic effects and can significantly slow AV nodal conduction. Effect: Chronic open-angle glaucoma and intraocular hypertension. |
Mechanistic genes:ADRB1, ADRB2 Metabolic genes Substrate:CYP2D6, CYP3A4, CYP3A5 |
|
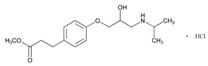
Name: CARVEDILOL IUPAC Name: 2-Propanol, 1-(9H-carbazol-4-yloxy)-3-[[2-(2-methoxyphenoxy)ethyl]amino]-, phosphate (salt), hydrate (2:2:1) Molecular Formula: C24H26N2O4 H3O4P ½H2O Molecular Weight: 513.48 Mechanism: Non-selective β-adrenergic blocker with α-adrenergic blocking activity. Available as a racemic mixture. Does not possess intrinsic sympathomimetic activity. Has calcium channel blocking activity at higher dose (30-fold the normal dose). In hypertensive patients, reduces cardiac output, exercise- or β-agonist-induced tachycardia, reflex orthostatic tachycardia, vasodilatation, peripheral vascular resistance (especially in standing position), renal vascular resistance, plasma renin activity, and increases levels of atrial natriuretic peptide. In congestive heart failure, decreases systemic blood pressure, pulmonary capillary wedge pressure, pulmonary artery pressure, heart rate, systemic vascular resistance, right arterial pressure, and increases stroke volume index and left ventricular ejection fraction. Effect: Mild-to-severe heart failure of ischemic or cardiomyopathic origin (usually in addition to standard therapy). Left ventricular dysfunction following myocardial infarction (clinically stable with LVEF ≤40%). Management of hypertension. |
Mechanistic genes:ADRA1A, ADRB1, ADRB2, GRK5, MMP2, NOX1 Metabolic genes Substrate:CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, UGT1A1, UGT1A4, UGT1A6, UGT2B7 Inhibitor:ABCB1 Transporter genes:ABCB1 |
|
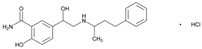
Name: ESMOLOL IUPAC Name: Benzenepropanoic acid, 4-[2-hydroxy-3-[(1-methylethyl)amino]propoxy]-, methyl ester, hydrochloride, (±)-; (±)-methyl p-[2-hydroxy-3-(isopropylamino)propoxy]hydrocinnamate hydrochloride Molecular Formula: C16H25NO4 HCl Molecular Weight: 331.83 Mechanism: A short-acting β1-selective adrenergic blocking agent. Competitively blocks response to β1-adrenergic stimulation with little or no effect on β2 receptors except at high doses, no intrinsic sympathomimetic activity, no membrane-stabilizing activity. Effect: Used in management of supraventricular tachyarrhythmias (e.g., atrial flutter and/or fibrillation, and sinus tachycardia). Used to prevent or treat increases in blood pressure associated with surgical events, including hypertensive crises (i.e., emergencies and urgencies). Treatment of non-compensatory sinus tachycardia. |
Mechanistic genes:
ADRB1
Metabolic genes Substrate: CYP2D6 |
|
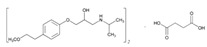
Name: LABETALOL IUPAC Name: Benzamide, 2-hydroxy-5-[1-hydroxy-2-[(1-methyl-3-phenylpropyl)amino]ethyl]-, monohydrochloride; 5-[1-hydroxy-2-[(1-methyl-3-phenylpropyl)amino]ethyl]salicylamide monohydrochloride Molecular Formula: C19H24N2O3 HCl Molecular Weight: 364.87 Mechanism: Blocks α-, β1-, and β2-adrenergic receptor sites. Elevated renins reduced. Effect: Treatment of mild to severe hypertension. Used intravenously for hypertensive emergencies. |
Mechanistic genes:ADRA, ADRB1, ADRB2 Metabolic genes Substrate:UGT1A1, UGT2B7 |
|
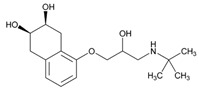
Name: METOPROLOL IUPAC Name: 2-Propanol, 1-[4-(2-methoxyethyl)phenoxy]-3-[(1-methylethyl)amino]-, (±)-, butanedioate (2:1); (±)-1-(isopropylamino)-3-[p-(2-methoxyethyl)phenoxy]-2-propanol succinate (2:1) Molecular Formula: (C15H25NO3)2 C4H6O4 Molecular Weight: 652.82 Mechanism: Competitively blocks β1 receptors, with little or no effect on β2 receptors at doses <100 mg. Effect: Treatment of angina pectoris, hypertension, or hemodynamically stable acute myocardial infarction. Treatment of angina pectoris or hypertension. To reduce mortality/hospitalization in heart failure patients already receiving ACEIs, diuretics, and/or digoxin. |
Mechanistic genes:ADRB1, ADRA2C, ACE, GRK5, KCNH2 Metabolic genes Substrate:CYP2C19, CYP2D6 Inhibitor:CYP2D6 Transporter genes:ABCB1 |
|
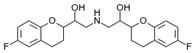
Name: NADOLOL IUPAC Name: 2,3-Naphthalenediol, 5-[3-[(1,1-dimethylethyl)amino]-2-hydroxypropoxy]-1,2,3,4-tetrahydro-, cis-; 1-(tert-butylamino)-3-[(5,6,7,8-tetrahydro-cis-6,7-dihydroxy-1-naphthyl)oxy]-2-propanol Molecular Formula: C17H27NO4 Molecular Weight: 309.40 Mechanism: Competitively blocks response to β1- and β2-adrenergic stimulation. Effect: Treatment of hypertension and angina pectoris. Prophylaxis of migraine headaches. |
Mechanistic genes:ADRB1, ADRB2, ADRB3 Transporter genes:ABCB1 Pleiotropic genes:IL10, IL12B |
|
Name: NEBIVOLOL IUPAC Name: 2-H-1-benzopyran-2-methanol, α,α’-[iminobis(methylene)bis[6-fluoro-3,4-dihydro-; α,α’-(iminodimethylene)bis[6-fluoro-2-chromanmethanol] Molecular Formula: C22H25F2NO4 Molecular Weight: 405.44 Mechanism: Highly-selective inhibitor of β1-adrenergic receptors. Nebivolol, unlike other β-blockers, also produces an endothelium-derived nitric oxide-dependent vasodilation resulting in a reduction in systemic vascular resistance. Effect: Treatment of hypertension, alone or in combination with other agents. |
Mechanistic genes:ADRB1 Metabolic genes Substrate:CYP2D6, UGTs Inducer:NOS3 |
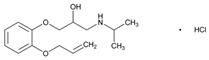
|
Name: OXPRENOLOL IUPAC Name: 2-Propanol, 1-(o-allyloxyphenoxy)-3-isopropylamino-, hydrochloride Molecular Formula: C15H23NO3 HCl Molecular Weight: 301.81 Mechanism: A competitive and non-selective antagonist of β-adrenergic receptors. Antagonizes catecholamine-induced tachycardia, thus decreasing cardiac output. Inhibits renin release by kidneys and inhibits vasomotor centers. Effect: Mild or moderate hypertension. |
Mechanistic genes:ADRB1, ADRB2, ADRB3 Metabolic genes Inhibitor:CYP2D6 |
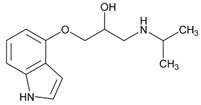
|
Name: PINDOLOL IUPAC Name: 2-Propanol, 1-(1H-indol-4-yloxy)-3-(1-methylethyl)amino-; 1-(indol-4-yloxy)-3-(isopropylamino)-2-propanol Molecular Formula: C14H20N2O2 Molecular Weight: 248.32 Mechanism: Blocks both β1 and β2 receptors and has mild intrinsic sympathomimetic activity. Has negative inotropic and chronotropic effects and can significantly slow AV nodal conduction. Augmentative action of antidepressants thought to be mediated via serotonin 1A autoreceptor antagonism. Effect: Treatment of hypertension, alone or in combination with other agents. |
Mechanistic genes:ADRB1, ADRB2, ADRB3, GRK5, HTR1A, HTR1B Metabolic genes Substrate:CYP2D6 Inhibitor:CYP2D6 |
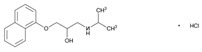
|
Name: PROPRANOLOL IUPAC Name: 2-Propanol, 1-[(1-methylethyl)amino]-3-(1-naphthalenyloxy)-, hydrochloride, (±)-; (±)-1-(isopropylamino)-3-(1-naphthyloxy)-2-propanol hydrochloride Molecular Formula: C16H21NO2 HCl Molecular Weight: 295.80 Mechanism: Competitively blocks response to β1- and β2-adrenergic stimulation, which results in decreases in heart rate, myocardial contractility, blood pressure, and myocardial oxygen demand. Reduces portal pressure by producing splanchnic vasoconstriction (β2-effect) thereby reducing portal blood flow. Effect: Management of hypertension. Angina pectoris. Pheochromocytoma. Essential tremor. Supraventricular arrhythmias (such as atrial fibrillation and flutter, AV nodal re-entrant tachycardias), ventricular tachycardias (catecholamine-induced arrhythmias, digoxin toxicity). Prevention of myocardial infarction. Migraine headache prophylaxis. Symptomatic treatment of hypertrophic subaortic stenosis (hypertrophic obstructive cardiomyopathy). |
Mechanistic genes:ADRB1, ADRB2, ADRB3, ALOX5, CFTR, COMT, FOS, GNAS, GRK5, HTR1B, HTR3B, KCNE2; KCNH2, KCNQ1, NPY, PPARGC1A, PTGS2 Metabolic genes Substrate:CYP1A1, CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, UGT2B7 Inhibitor:ABCB1, CYP1A2, CYP2D6 Transporter genes:ABCB1 Pleiotropic genes:IL1B, TNF |
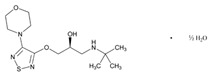
|
Name: TIMOLOL IUPAC Name: 2-Propanol, 1-[(1,1-dimethylethyl)amino]-3-[[4-(4-morpholinyl)-1,2,5-thiadiazol-3-yl]oxy]-, hemihydrate, (S)-; (S)-1-(tert-butylamino)-3-[(4-morpholino-1,2,5-thiadiazol-3-yl)oxy]-2-propanol hemihydrate Molecular Formula: C13H24N4O3S ½H2O Molecular Weight: 325.43 Mechanism: Blocks both β1- and β2-adrenergic receptors, reduces intraocular pressure by reducing aqueous humor production or possibly outflow. Reduces blood pressure by blocking adrenergic receptors and decreasing sympathetic outflow, produces negative chronotropic and inotropic activity through unknown mechanism. Effect: Treatment of elevated intraocular pressure such as glaucoma or ocular hypertension. Treatment of hypertension and angina, to reduce mortality following myocardial infarction. Prophylaxis of migraine. |
Mechanistic genes:ADRB1, GNAS Metabolic genes Substrate:CYP2C19, CYP2D6 Inhibitor:CYP2D6 |
| Calcium-Channel Blockers | ||
| Drug | Properties | Pharmacogenetics |
|
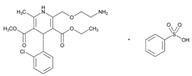
Name: AMLODIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 2-[(2-aminoethoxy)methyl]-4-(2-chlorophenyl)-1,4-dihydro-6-methyl-, 3-ethyl 5-methyl ester, (±)-, monobenzenesulfonate; 3-ethyl 5-methyl (±)-2-[(2-aminoethoxy)methyl]-4-(o-chlorophenyl)-1,4-dihydro-6-methyl-3,5-pyridinedicarboxylate, monobenzenesulfonate Molecular Formula: C20H25ClN2O5 C6H6O3S Molecular Weight: 567.05 Mechanism: Inhibits calcium ion from entering the voltage-sensitive channels of vascular smooth muscle and myocardium during depolarization, producing a relaxation of coronary vascular smooth muscle and coronary vasodilation. Increases myocardial oxygen delivery in vasospastic angina. Effect: Treatment of hypertension, symptomatic chronic stable angina, vasospastic (Prinzmetal’s) angina. Prevention of hospitalization due to angina with documented coronary artery disease. |
Mechanistic genes:ADD1, AGT, CACNs, NPPA Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP1A1, CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2D6, CYP3A4, CYP3A5 Transporter genes:ABCB1 |
|
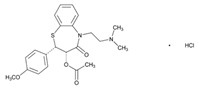
Name: DILTIAZEM IUPAC Name: 1,5-Benzothiazepin-4(5H)-one, 3-(acetyloxy)-5-[2-(dimethylamino)ethyl]-2,3-dihydro-2-(4-methoxyphenyl)-, monohydrochloride, (+)-cis-; (+)-5-[2-(dimethylamino)ethyl]-cis-2,3-dihydro-3-hydroxy-2-(p-methoxyphenyl)-1,5-benzothiazepin-4(5H)-one acetate (ester) monohydrochloride Molecular Formula: C22H26N2O4S HCl Molecular Weight: 450.98 Mechanism: Inhibits calcium ions from entering the “slow channels” or select voltage-sensitive areas of vascular smooth muscle and myocardium during depolarization, producing a relaxation of coronary vascular smooth muscle and coronary vasodilation. Increases myocardial oxygen delivery in vasospastic angina. Effect: Essential hypertension, chronic stable angina or angina from coronary artery spasm. Temporary control of rapid ventricular rate in atrial fibrillation or flutter. Management of supraventricular tachyarrhythmias, including rapid conversion to sinus rhythm of paroxysmal supraventricular tachycardias (e.g., those associated with Wolff–Parkinson–White or Lown–Ganong–Levine syndrome). |
Mechanistic genes:CACNA Metabolic genes Substrate:CYB5s, CYP2C8, CYP2C9, CYP2D6, CYP3A4, CYP3A5 Inhibitor:ABCB1, CYB5s, CYP2C9, CYP2D6, CYP3A4, CYP3A5 Transporter genes:ABCB1, SLCO1B1 Pleiotropic genes:IL12B |
|
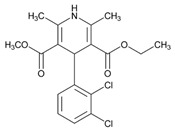
Name: FELODIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid 4-(2,3-dichlorophenyl)-1,4-dihydro-2,6-dimethyl-, ethyl methyl ester, (±)-; (±)-ethyl methyl 4-(2,3-dichlorophenyl)-1,4-dihydro-2,6-dimethyl-3,5-pyridinedicarboxylate Molecular Formula: C18H19Cl2NO4 Molecular Weight: 384.25 Mechanism: Inhibits calcium ions from entering the “slow channels” or select voltage-sensitive areas of vascular smooth muscle and myocardium during depolarization, producing a relaxation of coronary vascular smooth muscle and coronary vasodilation. Increases myocardial oxygen delivery in vasospastic angina. Effect: Treatment of hypertension. |
Mechanistic genes:CACNA1C, NR1I2 Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP2C8, CYP2C9, CYP2D6, CYP3A4, CYP3A5 |
|
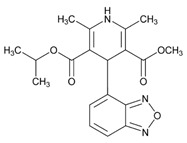
Name: ISRADIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 4-(4-benzofurazanyl)-1,4-dihydro-2,6-dimethyl-, methyl 1-methylethyl ester, (±)-; isopropyl methyl (±)-4-(4-benzofurazanyl)-1,4-dihydro-2,6-dimethyl-3,5-pyridinedicarboxylate Molecular Formula: C19H21N3O5 Molecular Weight: 371.39 Mechanism: Inhibits transmembrane influx of extracellular calcium ions across membranes of myocardial cells and vascular smooth muscle cells, without changing serum calcium concentrations. Increases myocardial oxygen delivery in vasospastic angina. Effect: Management of hypertension (alone or in combination with other classes of antihypertensive agents). |
Mechanistic genes:CACNA1C, NR1I2 Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:CYP3A4, CYP3A5 |
|
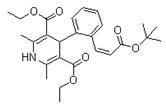
Name: LACIDIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 4-[2-[3-(1,1-dimethylethoxy)-3-oxo-1-propenyl]phenyl]-1,4-dihydro-2,6-dimethyl-, diethyl ester, I-; 4-I(E)-2-carboxyvinyl]-phenyl]-1,4-dihydro-2,6-dimethyl-3,5-pyridinedicarboxylic acid, 4-tert-butyl diethyl ester Molecular Formula: C26H33NO6 Molecular Weight: 455.54 Mechanism: A specific and potent calcium antagonist with predominant selectivity for calcium channels in vascular smooth muscle. Its main action is to dilate peripheral arterioles, reducing peripheral vascular resistance and lowering blood pressure. Effect: Indicated for treatment of hypertension either alone or in combination with other antihypertensive agents, including β-adrenoceptor antagonists, diuretics, and ACEIs. |
Mechanistic genes:CACN Metabolic genes Substrate: CYP3A4, CYP35 Inhibitor: CYP3A4, CYP35 |
|
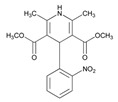
Name: NICARDIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 1,4-dihydro-2,6-dimethyl-4-(3-nitrophenyl)-, methyl 2-[methyl(phenylmethyl)amino]ethyl ester, monohydrochloride; 2-(benzylmethylamino)ethyl methyl 1,4-dihydro-2,6-dimethyl-4-(m-nitrophenyl)-3,5-pyridinedicarboxylate monohydrochloride Molecular Formula: C26H29N3O6 HCl Molecular Weight: 515.99 Mechanism: Inhibits calcium ions from entering “slow channels” or select voltage-sensitive areas of vascular smooth muscle and myocardium during depolarization, producing relaxation of coronary vascular smooth muscle and coronary vasodilation. Increases myocardial oxygen delivery in vasospastic angina. Effect: Chronic stable angina. Management of hypertension. |
Mechanistic genes:CACNA1C, CACNA1D, CACNA2D1, CACNB2 Metabolic genes Substrate:CYP1A1, CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2D6, CYP2E1, CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP1A1, CYP2A6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5 Inducer:CYP1A2 Transporter genes:ABCB1 |
|
Name: NIFEDIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 1,4-dihydro-2,6-dimethyl-4-(2-nitrophenyl)-, dimethyl ester; dimethyl 1,4-dihydro-2,6-dimethyl-4-(o-nitrophenyl)-3,5-pyridinedicarboxylate Molecular Formula: C17H18N2O6 Molecular Weight: 346.33 Mechanism: Inhibits calcium ions from entering “slow channels” or select voltage-sensitive areas of vascular smooth muscle and myocardium during depolarization, producing relaxation of coronary vascular smooth muscle and coronary vasodilation. Increases myocardial oxygen delivery in vasospastic angina. Effect: Angina and hypertension. Pulmonary hypertension. |
Mechanistic genes:ACE1, ACE2, CACNA1C, DRD2, FOS, MMP2, SCN5A Metabolic genes Substrate:CYP2D6, CYP2C8, CYP11B2, CYP3A4, CYP3A5, POR Inhibitor:ABCB1, CYP1A2, CYP2C9, CYP2D6, CYP2E1, CYP3A4, CYP3A5 Transporter genes:ABCB1, ABCC2, ABCC3, SLC14A2 Pleiotropic genes:TP53 |
|
Name: NIMODIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 1,4-dihydro-2,6-dimethyl-4-(3-nitrophenyl)-, 2-methoxyethyl 1-methylethyl ester; isopropyl 2-methoxyethyl 1,4-dihydro-2,6-dimethyl-4-(m-nitrophenyl)-3,5-pyridinedicarboxylate Molecular Formula: C21H26N2O7 Molecular Weight: 418.44 Mechanism: Animal studies indicate that nimodipine has greater effect on cerebral arterials than other arterials; this increased specificity may be due to increased lipophilicity and cerebral distribution of the drug as compared to nifedipine. Inhibits calcium ions from entering “slow channels” or select voltage-sensitive areas of vascular smooth muscle and myocardium during depolarization. Effect: Spasm following subarachnoid hemorrhage from ruptured intracranial aneurysms regardless of patient’s postictus neurological condition. |
Mechanistic genes:CACNA1C, DRD2 Metabolic genes Substrate:CYP3A4, CYP35 Pleiotropic genes:APP |
|
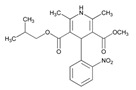
Name: NISOLDIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 1,4-dihydro-2,6-dimethyl-4-(2-nitrophenyl)-, methyl 2-methylpropyl ester, (±)-; (±)-isobutyl methyl 1,4-dihydro-2,6-dimethyl-4-(o-nitrophenyl)-3,5-pyridinedicarboxylate Molecular Formula: C20H24N2O6 Molecular Weight: 388.41 Mechanism: As a dihydropyridine calcium-channel blocker, structurally similar to nifedipine, nisoldipine impedes movement of calcium ions into vascular smooth muscle and cardiac muscle. Dihydropyridines are potent vasodilators and not as likely to suppress cardiac contractility and slow cardiac conduction as other calcium antagonists such as verapamil and diltiazem. Nisoldipine is 5–10-fold as potent vasodilator as nifedipine. Effect: Management of hypertension, alone or in combination with other antihypertensive agents. |
Mechanistic genes:CACNA1C Metabolic genes Substrate:CYP34, CYP3A5 Inhibitor:ABCB1, CYP1A2, CYP3A4, CYP3A5 |
|
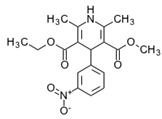
Name: NITRENDIPINE IUPAC Name: 3,5-Pyridinedicarboxylic acid, 1,4-dihydro-2,6-dimethyl-4-(3-nitrophenyl)-, ethyl methyl ester, (±)-; (±)-ethyl methyl 1,4dihydro-2,6-dimethyl-4-(m-nitrophenyl)-3,5-pyridinedicarboxylate Molecular Formula: C18H20N2O6 Molecular Weight: 360.36 Mechanism: Dihydropyridine calcium channel-blocking agent with actions similar to nifedipine. Effect: Management of hypertension. |
Mechanistic genes:CACNA1C, CACNG1 Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP3A4, CYP3A5 Transporter genes:ABCG2 |
|
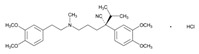
Name: VERAPAMIL IUPAC Name: Benzeneacetonitrile, α-[3-[[2-(3,4-dimethoxyphenyl)ethyl]methylamino]propyl]-3,4-dimethoxy-α-(1-methylethyl)-, monohydrochloride, (±)-; (±)-5-[(3,4-dimethoxyphenethyl)methylamino]-2-(3,4-dimethoxyphenyl)-2-isopropylvaleronitrile monohydrochloride Molecular Formula: C27H38N2O4 HCl Molecular Weight: 491.06 Mechanism: Inhibits calcium ions from entering “slow channels” or select voltage-sensitive areas of vascular smooth muscle and myocardium during depolarization. Produces relaxation of coronary vascular smooth muscle and coronary vasodilation. Increases myocardial oxygen delivery in vasospastic angina. Slows automaticity and conduction of the AV node. Effect: Orally for treatment of angina pectoris (vasospastic, chronic stable, and unstable) and hypertension. I.V. for supraventricular tachyarrhythmias (PSVT, atrial fibrillation, and atrial flutter). |
Mechanistic genes:ADRB1, ADRB2, CACNA1C, CACNs, CFTR, KCNMB1, LDLR, NOS1AP, RET, TGFB1 Metabolic genes Substrate:CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C18, CYP2C19, CYP2E1, CYP2J2, CYP3A4, CYP3A5, CYP3A7, SOD2 Inhibitor:ABCB1, ABCC1, ABCC2, CYP1A2, CYP2C8, CYP2C9, CYP2D6, CYP3A4, CYP3A5, CYP3A7 Inducer:ABCB1 Transporter genes:ABCB1, ABCC3, SLCO1B1 Pleiotropic genes:TNF |
| Cardiotonic Agents | ||
| Drug | Properties | Pharmacogenetics |
|
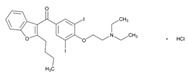
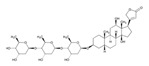
Name: AMIODARONE IUPAC Name: Methanone, (2-butyl-3-benzofuranyl)[4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl]-; 2-butyl-3-benzofuranyl 4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl ketone Molecular Formula: C25H29I2NO3 Molecular Weight: 645.31 Mechanism: Inhibits adrenergic stimulation (α- and β-blocking properties), affects sodium, potassium and calcium channels, and prolongs the action potential and refractory period in myocardial tissue. Decreases AV conduction and sinus node function. Effect: Management of recurrent ventricular fibrillation or hemodynamically-unstable ventricular tachycardia. |
Mechanistic genes:ABL1, ACOX1, ADRA2A, ADRB1, ADRB2, CHRM2, FABP1, FMO1, FOS, ICAM1, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1, PSEN1, SCN5A Metabolic genes Substrate:CYP1A1, CYP1A2, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2J2, CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5 Inducer:CYP1A2 Transporter genes:ABCB1, ABCC6, ABCC8, SLC5A5 |
|
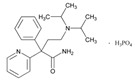
Name: DIGOXIN IUPAC Name: Card-20(22)-enolide, 3-[(O-2,6-dideoxy-β-D-ribo-hexopyranosyl-(1→4)-O-2,6-dideoxy-β-D-ribo-hexopyranosyl-(1→4)-2,6-dideoxy-β-D-ribo-hexopyranosyl)oxy]-12,14-dihydroxy-, (3β,5β,12β)-; 3β-[(O-2,6-dideoxy-β-D-ribo-hexopyranosyl-(1→4)-O-2,6-dideoxy-β-D-ribo-hexopyranosyl-(1→4)-2,6-dideoxy-β-D-ribo-hexopyranosyl)oxy]-12β,14-dihydroxy-5β-card-20(22)-enolide Molecular Formula: C41H64O14 Molecular Weight: 780.94 Mechanism: Digoxin is a cardiac glycoside with positive inotropic effects. In congestive heart failure it inhibits the sodium/potassium ATPase pump which acts to increase the intracellular sodium-calcium exchange to increase intracellular calcium, leading to increased contractility. In supraventricular arrhythmias: Direct suppression of the AV node conduction to increase effective refractory period and decrease conduction velocity; positive inotropic effect, enhanced vagal tone, and decreased ventricular rate to fast atrial arrhythmias. Atrial fibrillation may decrease sensitivity and increase tolerance to higher serum digoxin concentrations. Effect: Digitalization and maintenance therapy. Used principally in the prophylactic management and treatment of congestive heart failure and to control the ventricular rate in supraventricular tachyarrhythmias (e.g., atrial fibrillation or flutter). Used to improve left ventricular function in cardiogenic shock and atrial fibrillation or flutter with rapid ventricular rate. May be useful, especially in conjunction with a β-adrenergic blocking agent, in the treatment of angina pectoris when cardiomegaly and congestive heart failure are present. |
Mechanistic genes:ATP1A1 Metabolic genes Substrate:CYP3A4, CYP3A5 Transporter genes:ABCB1, ABCB11, ABCG2, SLCO1B3 |
|
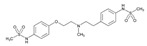
Name: DISOPYRAMIDE IUPAC Name: 2-Pyridineacetamide, α-[2-[bis(1-methylethyl)amino]ethyl]-α-phenyl-, (±)-, phosphate (1:1); (±)-α-[2-(diisopropylamino)ethyl]-α-phenyl-2-pyridineacetamide phosphate (1:1) Molecular Formula: C21H29N3O H3PO4 Molecular Weight: 437.47 Mechanism: Decreases myocardial excitability and conduction velocity. Reduces disparity in refractory period between normal and infarcted myocardium. Possesses anticholinergic, peripheral vasoconstrictive, and negative inotropic effects. Effect: Suppression and prevention of unifocal and multifocal ventricular premature complexes, coupled ventricular premature complexes and/or paroxysmal ventricular tachycardia in primary arrhythmias or arrhythmias secondary to coronary artery disease. |
Mechanistic genes:ADRB1, ADRB2, CHRM2, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1 Metabolic genes Substrate:CYP2D6, CYP3A4, CYP3A5 Inhibitor:CYP1A1, CYP1A2, CYP2C19, CYP3A4, CYP3A5 Transporter genes:ABCC8 |
|
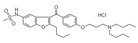
Name: DOFETILIDE IUPAC Name: Methanesulfonamide, N-[4-[2-[methyl[2-[4-[(methylsulfonyl)amino]phenoxy]ethyl]amino]ethyl]phenyl]-; β-[(p-methanesulfonamidophenethyl)methylamino]methanesulfono-p-phenetidide Molecular Formula: C19H27N3O5S2 Molecular Weight: 441.56 Mechanism: Class III antiarrhythmic agent. Blockade of the cardiac ion channel carrying the rapid component of the delayed rectifier potassium current. It increases the monophasic action potential duration due to delayed repolarization. The increase in the QT interval is a function of prolongation of both effective and functional refractory periods in the His–Purkinje system and the ventricles. Effect: Used for the maintenance of normal sinus rhythm in patients with atrial fibrillation/flutter of more than 1 week duration who have been converted to normal sinus rhythm. Additionally used for the conversion of atrial fibrillation and atrial flutter to normal sinus rhythm. |
Mechanistic genes:ADRA2A, ADRB1, CHRM2, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1 Metabolic genes Substrate:CYP2D6, CYP3A4, CYP3A5 Transporter genes:ABCC8 |
|
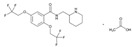
Name: DRONEDARONE IUPAC Name: N-[2-butyl-3-[4-[3-(dibutylamino)propoxy]benzoyl]-1-benzofuran-5-yl] methanesulfonamide, hydrochloride Molecular Formula: C31H44N2O5S HCl Molecular Weight: 593.22 Mechanism: Dronedarone has antiarrhythmic properties belonging to all four antiarrhythmic (Vaughan-Williams) classes, but the contribution of each of these activities to the clinical effect is unknown. Inhibits sodium (INa) and potassium (Ikr, IkS, Ik1, and Ik-ACh) channels resulting in prolongation of the action potential and refractory period in myocardial tissue without reverse-use-dependent effects. Decreases AV conduction and sinus node function through inhibition of calcium (ICa-L) channels and β1-receptor blocking activity. Similar to amiodarone, dronedarone also inhibits α1-receptor-mediated increases in blood pressure. Effect: Indicated to reduce the risk of hospitalization related to paroxysmal or persistent atrial fibrillation (AF) or atrial flutter (AFl) in patients with a recent episode of AF/AFl and associated cardiovascular risk factors (e.g., age >70 years, hypertension, diabetes, prior cerebrovascular accident, left atrial diameter ≥50 mm or left ventricular ejection fraction <40%), who are in normal sinus rhythm or will be cardioverted. |
Mechanistic genes:KCNA5, KCNE2, KCNH2 Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP2C9, CYP2D6, CYP3A4, CYP3A5 |
|
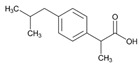
Name: FLECAINIDE IUPAC Name: Benzamide, N-(2-piperidinylmethyl)-2,5-bis(2,2,2-trifluoroethoxy)-, monoacetate Molecular Formula: C17H20F6N2O3 C2H4O2 Molecular Weight: 474.39 Mechanism: Flecainide is a local anesthetic-type class Ic antiarrhythmic agent. Slows conduction in cardiac tissue by altering transport of ions across cell membranes. Causes slight prolongation of refractory periods. Decreases rate of rise of action potential without affecting duration. Increases electrical stimulation threshold of ventricle, the His–Purkinje system. Possesses local anesthetic and moderate negative inotropic effects. Effect: Prevention and suppression of documented life-threatening ventricular arrhythmias (e.g., sustained ventricular tachycardia). Control of symptomatic, disabling supraventricular tachycardias in patients without structural heart disease where other agents fail. |
Mechanistic genes:ADRA2A, CHRM2, FABP1, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1, SCN5A Metabolic genes Substrate:CYP1A2, CYP2D6 Inhibitor:CYP2D6 Transporter genes:ABCC8 |
|
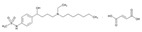
Name: IBUPROFEN IUPAC Name: Benzeneacetic acid, α-methyl-4-(2-methylpropyl), (±)-; (±)-p-isobutylhydratropic acid; (3)(±)-2-(p-isobutylphenyl)propionic acid Molecular Formula: C13H18O2 Molecular Weight: 206.28 Mechanism: Inhibits prostaglandin synthesis by decreasing activity of enzyme cyclooxygenase (COX/PTGS), which results in decreased formation of prostaglandin precursors. Effect: Inflammatory diseases and rheumatoid disorders including juvenile rheumatoid arthritis, mild-to-moderate pain, fever, dysmenorrhea. Ibuprofen lysine is for use in premature infants weighing between 500 and 1500 g and who are ≤32 weeks gestational age to induce closure of clinically significant patent ductus arteriosus (PDA) when usual treatments are ineffective. Management of pain and swelling. |
Mechanistic genes:ACSL1, AGT, BCAR1, CCND1, CNR1, ERBB4, FOS, ICAM1, IFNG, IGF1, LTA, MMP3, NOS3, NQO1, NR3C1, PPARA, PPARG, PTGER3, PTGES, PTGIS, PTGS1, PTGS2, RB1, REN, SCARB1, SNCA, TBX21, TBXAS1, VCAM1, VEGFA Metabolic genes Substrate:CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, CYP19A1, GSTT1, SOD2, UGT1A1, UGT1A3, UGT1A7, UGT1A9, UGT1A10, UGT2B7 Inhibitor:ACACA, CYP2C9, DDC, PTGS2, SLC5A8, SULT1A1, UGT2B15 Inducer:CYP19A1 Transporter genes:SLC15A1, SLC22A1, SLC22A6, SLC22A7, SLC22A8 Pleiotropic genes:APP, IL1B, IL1RN, IL4, IL6, IL10, TNF |
|
Name: IBUTILIDE IUPAC Name: Methanesulfonamide, N-[4-[4-(ethylheptylamino)-1-hydroxybutyl]phenyl]-, (±)-, (E)-2-butenedioate (2:1); (±)-4′-[4-(ethylheptylamino)-1-hydroxybutyl]methanesulfonanilide fumarate (2:1) Molecular Formula: (C20H36N2O3S)2 C4H4O4 Molecular Weight: 885.23 Mechanism: Prolongs action potential duration and effective refractory period in both atrial and ventricular cardiac tissue. Delays repolarization by activating a slow, predominantly sodium, inward current. Produces dose-related prolongation of QT interval, thought to be associated with antiarrhythmic activity. Negligible effects on heart rate, cardiac contractility, or blood pressure. Lacks β-adrenergic-blocking activity. Effect: Used for rapid conversion of recent-onset atrial flutter or fibrillation to sinus rhythm. |
Mechanistic genes:ADRA2A, CACNA1C, CHRM2, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1 Metabolic genes Substrate:CYP2D6 Transporter genes:ABCB1, ABCC8 |
|
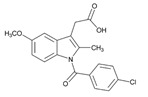
Name: INDOMETHACIN IUPAC Name: 1H-indole-3-acetic acid, 1-(4-chlorobenzoyl)-5-methoxy-2-methyl-; 1-(p-chlorobenzoyl)-5-methoxy-2-methylindole-3-acetic acid Molecular Formula: C19H16ClNO4 Molecular Weight: 357.79 Mechanism: Inhibits cyclooxygenase-1 (COX-1/PTGS1) and COX-2/PTGS2. Exhibits anti-inflammatory, analgesic, and antipyretic activity. Permits closure of ductus arteriosus in premature neonates by inhibiting prostaglandin synthesis. Effect: Symptomatic treatment of osteoarthritis, rheumatoid arthritis, and ankylosing spondylitis. Symptomatic relief of acute gout and acute painful shoulder (i.e., bursitis and/or tendinitis). Treatment of patent ductus arteriosus in premature neonates. |
Mechanistic genes:AGTR1, BCAR1, CAT, CBR1, CBS, CCND1, CDK2, CDK4, COL1A1, DIO2, EDN1, EGFR, FOS, G6PD, GNAS, ICAM1, IFNG, LEP, LPL, LTA, MAPT, MMP2, MMP3, NOS3, NPPA, NPY, PARK2, PDGFRA, PDGFRB, PPARA, PPARD, PPARG, PTGER2, PTGES, PTGIS, PTGS1, PTGS2, RB1, TBXAS1, TGFB1, TGFBR1, THBD, TLR, UCP2, VEGFA, XDH Metabolic genes Substrate:CYP1A1, CYP1A2, CYP1B1, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, CYP7A1, CYP19A1, CYP27A1, GSTM1, GSTT1, SOD2, UGT1A1, UGT1A3, UGT1A4, UGT1A7, UGT1A9, UGT1A10, UGT2B7 Inhibitor: CYP2C9, CYP2C19, PTGS1, PTGS2, SLC22A7, SULT1A1 Transporter genes:ABCB1, ABCC1, ABCC2, ABCC3, ABCC6, ABCG2, SLC10A1, SLC19A1, SLC22A1, SLC22A2, SLC22A6, SLC22A7, SLC22A8 Pleiotropic genes:APC, APP, IL1B, IL1RN, IL2, IL4, IL6, IL10, IL12B, TNF, TNFRSF1A, TP53 |
|
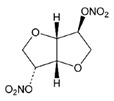
Name: ISOSORBIDE DINITRATE IUPAC Name: D-glucitol, 1,4:3,6-dianhydro-, dinitrate Molecular Formula: C6H8N2O8 Molecular Weight: 236.14 Mechanism: Stimulation of intracellular cyclic GMP results in vascular smooth muscle relaxation of both arterial and venous vasculature. Effect: Acute relief of angina pectoris, prophylactic management in situations likely to provoke angina attacks, and long-term prophylactic management of angina pectoris. Additionally used to relieve pain, dysphagia, and spasm in esophageal spasm with gastroesophageal reflux. |
Mechanistic genes:ALDH3A1, EDN1, NOS3 Metabolic genes Substrate:CYP3A4, CYP3A5, CYP11B2 |
|
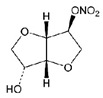
Name: ISOSORBIDE MONONITRATE IUPAC Name: D-glucitol, 1,4:3,6-dianhydro-, 5-nitrate; 1,4:3,6-dianhydro-D-glucitol 5-nitrate Molecular Formula: C6H9NO6 Molecular Weight: 191.14 Mechanism: Decreases preload as measured by pulmonary capillary wedge pressure and left ventricular end diastolic volume and pressure. This effect improves congestive symptoms in heart failure and improves myocardial perfusion gradient in coronary artery disease. Effect: Long-acting metabolite of vasodilator isosorbide dinitrate used for prophylactic treatment of angina pectoris. |
Mechanistic genes:NOS3 Metabolic genes Substrate:CYP3A4, CYP3A5, CYP11B2 |
|
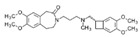
Name: IVABRADINE IUPAC Name: 3-[3-[[[(7S)-3,4-dimethoxybicyclo[4.2.0]octa-1,3,4,5-tetrahydro-7,8-dimethoxy-2H-3-benzazepin-2-one Molecular Formula: C27H36N2O5 Molecular Weight: 468.59 Mechanism: Ivabradine is a pure heart rate-lowering agent, acting by selective and specific inhibition of the cardiac pacemaker If current that controls the spontaneous diastolic depolarization in the sinus node and regulates heart rate. The cardiac effects are specific to the sinus node with no effect on intra-atrial, atrioventricular or intraventricular conduction times, nor on myocardial contractility or ventricular repolarization. Effect: Symptomatic treatment of chronic stable angina pectoris in patients with normal sinus rhythm, who have a contraindication or intolerance for β-blockers. |
Mechanistic genes:HCN1, HCN4 Metabolic genes Substrate:CYP3A4, CYP3A5 |
|
Name: LIDOCAINE IUPAC Name: Acetamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)-; (2) 2-(diethylamino)-2′,6′-acetoxylidide Molecular Formula: C14H22N2O Molecular Weight: 234.34 Mechanism: Suppresses automaticity of conduction tissue, by increasing electrical stimulation threshold of ventricle, the His–Purkinje system, and spontaneous depolarization of ventricles during diastole by direct action on tissues. Effect: Local anesthetic and acute treatment of ventricular arrhythmias (such as from myocardial infarction or cardiac manipulation). |
Mechanistic genes:CHRM2, FOS, KCNE2, KCNH2, KCNJ11, KCNQ1, SCN5A, TRPV1, VEGFA Metabolic genes Substrate:CYP1A2, CYP2A6, CYP2B6, CYP2C9, CYP2D6, CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP1A2, CYP2D6, CYP3A4, CYP3A5 Transporter genes:ABCC8, SLC22A16 Pleiotropic genes:IL6 |
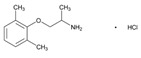
|
Name: MEXILETINE IUPAC Name: 2-Propanamine, 1-(2,6-dimethylphenoxy)-, hydrochloride, (±)-; (±)-1-methyl-2-(2,6-xylyloxy)ethylamine hydrochloride Molecular Formula: C11H17NO HCl Molecular Weight: 215.72 Mechanism: Inhibits inward sodium current, decreases rate of rise of phase 0, increases effective refractory period/action potential duration ratio. Effect: Management of serious ventricular arrhythmias. Suppression of premature ventricular contractions. |
Mechanistic genes:CHRM2, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1, SCN5A Metabolic genes Substrate:CYP1A2, CYP2D6, CYP3A4, CYP3A5 Inhibitor:CYP1A2 Transporter genes:ABCC8 |
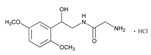
|
Name: MIDODRINE IUPAC Name: Acetamide, 2-amino-N-[2-(2,5-dimethoxyphenyl)-2-hydroxyethyl]-, monohydrochloride, (±)-; (±)-2-amino-N-(β-hydroxy-2,5-dimethoxyphenethyl)acetamide monohydrochloride Molecular Formula: C12H18N2O4 HCl Molecular Weight: 290.74 Mechanism: Midodrine forms an active metabolite, desglymidodrine, an α1 agonist. This agent increases arteriolar and venous tone resulting in a rise in standing, sitting, and supine systolic and diastolic blood pressure in orthostatic hypotension. Effect: Orphan drug: Treatment of symptomatic orthostatic hypotension. |
Mechanistic genes:ADRA1A, ADRA1B Metabolic genes Substrate:CYP2D6 |
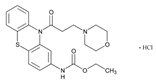
|
Name: MORICIZINE IUPAC Name: Carbamic acid, [10-[3-(4-morpholinyl)-l-oxopropyl]-10H-phenothiazin-2yl]-, ethyl ester, hydrochloride; ethyl 10-(3-morpholinopropionyl)phenothiazine-2-carbamate, hydrochloride Molecular Formula: C22H25N3O4S HCl Molecular Weight: 463.98 Mechanism: Reduces fast inward current carried by sodium ions, shortens phase I and phase II repolarization, resulting in decreased action potential duration and effective refractory period. Effect: Treatment of ventricular tachycardia and life-threatening ventricular arrhythmias. |
Mechanistic genes:SCN5A Metabolic genes Substrate:CYP3A4, CYP3A5 Inducer:CYP1A2, CYP3A4, CYP3A5 |
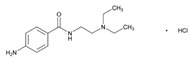
|
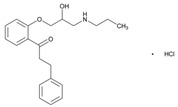
Name: PROCAINAMIDE IUPAC Name: Benzamide, 4-amino-N-[2-(diethylamino)ethyl]-, monohydrochloride; p-amino-N-[2-(diethylamino)ethyl]benzamide monohydrochloride Molecular Formula: C13H21N3O HCl Molecular Weight: 271.79 Mechanism: Decreases myocardial excitability and conduction velocity and may depress myocardial contractility, by increasing electrical stimulation threshold of ventricle, the His–Purkinje system and through direct cardiac effects. Effect: Treatment of ventricular tachycardia, premature ventricular contractions, paroxysmal atrial tachycardia, and atrial fibrillation. Prevention of recurrence of ventricular tachycardia, paroxysmal supraventricular tachycardia, atrial fibrillation or flutter. |
Mechanistic genes:CHRM2, KCNE1, KCNE2, KCNQ1, KCNH2, KCNJ11, SCN5A Metabolic genes Substrate:CYP2D6, CYP3A4, CYP3A5, NAT2 Transporter genes:ABCC8, SLC22A16 |
|
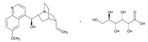
Name: PROPAFENONE IUPAC Name: 1-Propanone, 1-[2-[2-hydroxy-3-(propylamino)propoxy]phenyl]-3-phenyl-, hydrochloride; 2′-[2-hydroxy-3-(propylamino)propoxy]-3-phenylpropiophenone hydrochloride Molecular Formula: C21H27NO3 HCl Molecular Weight: 377.90 Mechanism: Possesses local anesthetic properties, blocks fast inward sodium current, and slows rate of increase of action potential. Prolongs effective refractory period, reduces spontaneous automaticity and exhibits some β-blockade activity. Effect: Treatment of life-threatening ventricular arrhythmias. Maintenance of normal sinus rhythm in symptomatic atrial fibrillation. |
Mechanistic genes:ADRB, CHRM2, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1 Metabolic genes Substrate:CYP1A1, CYP1A2, CYP2D6, CYP3A4, CYP3A5, UGTs Transporter genes:ABCC8 |
|
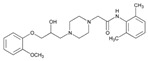
Name: QUINIDINE IUPAC Name: (8R,9S)-6′-Methoxycinchonan-9-ol Molecular Formula: C20H24N2O2 Molecular Weight: 324.42 Mechanism: Depresses phase 0 of action potential. Decreases myocardial excitability and conduction velocity, and myocardial contractility by decreasing sodium influx during depolarization and potassium efflux in repolarization. Additionally reduces calcium transport across cell membrane. Decreases conduction velocity in atria, ventricles, and the His–Purkinje system, and may decrease or cause no change in conduction velocity through the AV node. May suppress atrial fibrillation or flutter. May produce sinus tachycardia via its anticholinergic effects. Has direct negative inotropic effect, but therapeutic plasma concentrations of drug do not usually depress contractility in normal heart. May reduce peripheral resistance and blood pressure by blockade of α-adrenergic receptors and by effects on myocardial contractility. Acts principally as intraerythrocytic schizonticide (has little effect on sporozoites or pre-erythrocytic parasites). Gametocidal against Plasmodium vivax and P. malariae, but not P. falciparum. Effect: Prophylaxis after cardioversion of atrial fibrillation and/or flutter to maintain normal sinus rhythm. Prevent recurrence of paroxysmal supraventricular tachycardia, paroxysmal AV junctional rhythm, paroxysmal ventricular tachycardia, paroxysmal atrial fibrillation, and atrial or ventricular premature contractions. Has activity against Plasmodium falciparum malaria. |
Mechanistic genes:CHRM2, CHRNA1, FGB, G6PD, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1, LIPC, SCN5A, TNFRSF1A Metabolic genes Substrate:CYP1A2, CYP2C9, CYP2D6, CYP2E1, CYP3A4, CYP3A5, GSTM1, GSTP1 Inhibitor:ABCB1, CYP2C9, CYP2D6, CYP3A4, CYP3A5 Transporter genes:ABCB1, ABCC1, ABCC8 |
|
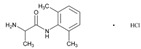
Name: RANOLAZINE IUPAC Name: 1-Piperazineacetamide, N-(2,6-dimethylphenyl)-4-[2-hydroxy-3-(2-methoxyphenoxy)propyl]-; (±)-N-(2,6-dimethylphenyl)-4-[2-hydroxy-3-(2-methoxyphenoxy)propyl]-1-piperazineacetamide Molecular Formula: C24H33N3O4 Molecular Weight: 427.54 Mechanism: Ranolazine exerts antianginal and anti-ischemic effects without changing hemodynamic parameters (heart rate or blood pressure). At therapeutic levels, ranolazine inhibits late phase of inward sodium channel (late INa) in ischemic cardiac myocytes during cardiac repolarization reducing intracellular sodium concentrations and thereby reducing calcium influx via Na+-Ca2+ exchange. Decreased intracellular calcium reduces ventricular tension and myocardial oxygen consumption. It is thought that ranolazine produces myocardial relaxation and reduces anginal symptoms through this mechanism although this is uncertain. At higher concentrations, ranolazine inhibits rapid delayed rectifier potassium current (IKr) thus prolonging ventricular action potential duration and subsequent prolongation of QT interval. Effect: Treatment of chronic angina. |
Mechanistic genes:SCN5A Metabolic genes Substrate:CYP2D6, CYP3A4, CYP3A5 Inhibitor:ABCB1, CYP2D6, CYP3A4, CYP3A5 Transporter genes:ABCB1 |
|
Name: TOCAINIDE IUPAC Name: Propanamide, 2-amino-N-(2,6-dimethylphenyl)-, hydrochloride, (±)-; (±)-amino-2′,6′-propionoxylidide hydrochloride Molecular Formula: C11H16N2O.HCl Molecular Weight: 228.72 Mechanism: Suppresses automaticity of conduction tissue, by increasing electrical stimulation threshold of ventricle, the His–Purkinje system, and spontaneous depolarization of ventricles during diastole by direct action on tissues. Blocks both initiation and conduction of nerve impulses by decreasing permeability to sodium ions of neuronal membrane, which results in inhibition of depolarization with resultant blockade of conduction. Effect: Suppression and prevention of symptomatic life-threatening ventricular arrhythmias. |
Mechanistic genes:CHRM2, KCNE1, KCNE2, KCNH2, KCNJ11, KCNQ1, SCN5A Inhibitor:CYP1A2 Transporter genes:ABCC8 |
|
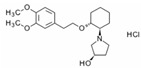
Name: VERNAKALANT IUPAC Name: (3R)-1-[(1R,2R)-2-[2-(3,4-dimethoxyphenyl)ethoxy]cyclo34enzazepineolidin-3-ol hydrochloride Molecular Formula: C20H31NO4 HCl Molecular Weight: 385.20 Mechanism: Vernakalant is an antiarrhythmic medicine that acts preferentially in the atria to prolong atrial refractoriness and to rate-dependently slow impulse conduction. These antifibrillatory actions on refractoriness and conduction are thought to suppress re-entry, and are potentiated in the atria during atrial fibrillation. The relative selectivity of vernakalant on atrial vs. ventricular refractoriness is postulated to result from the block of currents that are expressed in the atria, but not in the ventricles, as well as the unique electrophysiologic condition of the fibrillating atria. However, blockade of cationic currents, including hERG channels and cardiac voltage-dependent sodium channels present in the ventricles, has been documented. Effect: Rapid conversion of recent onset atrial fibrillation to sinus rhythm in adults (for non-surgery patients, atrial fibrillation ≤7 days duration; and for post-cardiac surgery patients, atrial fibrillation ≤3 days duration). |
Mechanistic genes:KCNA5, KCNH2 Metabolic genes Substrate:CYP2D6, UGTs |
| Diuretics | ||
| Drug | Properties | Pharmacogenetics |
|
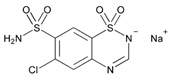
Name: CHLOROTHIAZIDE IUPAC Name: 2H-1,2,4-benzothiadiazine-7-sulfonamide, 6-chloro-, 1,1-dioxide, monosodium salt; 6-chloro-2H-1,2,4-benzothiadiazine-7-sulfonamide, 1,1-dioxide, monosodium salt Molecular Formula: C7H5ClN3NaO4S2 Molecular Weight: 317.71 Mechanism: Primary site of diuretic action appears to be the cortical diluting segment of the nephron. Enhances excretion of sodium, chloride, and water by interfering with the transport of sodium ions across the renal tubular epithelium. Enhances urinary excretion of potassium secondary to increased amount of sodium at distal tubular site of sodium–potassium exchange. Increases urinary bicarbonate excretion (although to a lesser extent than chloride excretion) but change in urinary pH is usually minimal (diuretic efficacy not affected by acid–base balance of patient). Increases calcium urinary excretion from a decrease in extracellular fluid volume, although calcium reabsorption in the nephron may be increased (also, slight or intermittent elevations in serum calcium concentration). Hypotensive activity in hypertensive patients (unknown mechanism; potential direct arteriolar dilation). Effect: Mild-to-moderate hypertension. Edema. |
Mechanistic genes:ADD1, ADRB1, ADRB2, ACE1, AGT, GNB3, NOS3, SCNN1G, WNK1 Metabolic genes Substrate:CYP11B2 |
|
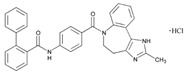
Name: CONIVAPTAN IUPAC Name: [1,1′-Biphenyl]-2-carboxamide, N-[4-[(4,5-dihydro-2-methylimidazo[4,535enzazepineazepin-6(1H)-yl)carbonyl]phenyl]-, monohydrochloride; 4′’-[(4,5-dihydro-2-methylimidazo[4,535enzazepineazepin-6(1H)-yl)carbonyl]-2-biphenylcarboxanilide monohydrochloride Molecular Formula: C32H26N4O2 HCl Molecular Weight: 535.04 Mechanism: Conivaptan is an arginine vasopressin (AVP) receptor antagonist with affinity for AVP receptor subtypes V1A and V2. The antidiuretic action of AVP is mediated through activation of the V2 receptor, which functions to regulate water and electrolyte balance at the level of the collecting ducts in the kidney. Antagonism of the V2 receptor by conivaptan promotes the excretion of free water (without loss of serum electrolytes) resulting in net fluid loss, increased urine output, decreased urine osmolality, and subsequent restoration of normal serum sodium levels. Blockade of vascular V1A receptors may cause splanchnic vasodilation, and thus hypotension or variceal bleeding in patients with cirrhosis (especially those with portal hypertension). Effect: Euvolemic and hypervolemic hyponatremia in hospitalized patients. |
Mechanistic genes:AVPR1A Metabolic genes Substrate:CYP3A4, CYP3A5 Inhibitor:CYP3A4, CYP3A5 |
|
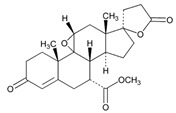
Name: EPLERENONE IUPAC Name: Pregn-4-ene-7,21-dicarboxylic acid, 9,11-epoxy-17-hydroxy-3-oxo-, γ-lactone, methyl ester, (7α,11α,17α)-; 9,11α-epoxy-17-hydroxy-3-oxo-17α-pregn-4-ene-7α,21-dicarboxylic acid, γ-lactone, methyl ester Molecular Formula: C24H30O6 Molecular Weight: 414.49 Mechanism: A relatively selective competitive mineralocorticoid (aldosterone) receptor antagonist. Binds selectively to mineralocorticoid receptors and has low (less than 1%) affinity for glucocorticoid, progesterone, and androgen receptors. It is a competitive antagonist of aldosterone at mineralocorticoid receptors in the kidney, myocardium, salivary gland, GI tract, brain, and vasculature, and has been shown to inhibit the physiologic effects of aldosterone in these organs. Some of the antihypertensive effects of eplerenone may be related to restoration of endothelial function by increasing the release of nitric oxide, which results in vasodilation. Has been shown to produce sustained increases in plasma renin and serum aldosterone concentrations, reflecting the inhibition of the negative feedback of aldosterone on renin secretion. Appears to have cardioprotective effects in congestive heart failure and left ventricular dysfunction following MI. The cardioprotective action mechanism appears to be related more to the ability of the drug to competitively inhibit the pathophysiologic effects of aldosterone on the myocardium than to its hypotensive effects. Eplerenone reduces coronary vascular inflammation, risk of subsequent development of interstitial myocardial and coronary perivascular fibrosis, cardiac hypertrophy, and/or ventricular remodeling. Effect: To reduce the risk of mortality following acute MI in clinically stable patients with left ventricular dysfunction (i.e., LVEF 40% or less) who have demonstrated clinical evidence of CHF. Used orally in the management of hypertension. May be used as monotherapy or in combination with other classes of antihypertensive agents (e.g., ACEIs, angiotensin II receptor antagonists, calcium-channel blocking agents, β-adrenergic blocking agents, and thiazide diuretics). |
Mechanistic genes:NOS3, NPPA, NR3C2 Metabolic genes Substrate:CYP3A4, CYP3A5, CYP11B2 |
|
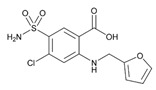
Name: FUROSEMIDE IUPAC Name: Benzoic acid, 5-(aminosulfonyl)-4-chloro-2-[(2-furanylmethyl)amino]-; 4-chloro-N-furfuryl-5-sulfamoylanthranilic acid Molecular Formula: C12H11ClN2O5S Molecular Weight: 330.74 Mechanism: Inhibits reabsorption of sodium and chloride in ascending loop of Henle and distal renal tubule, interfering with chloride-binding cotransport system, thus causing increased excretion of water, sodium, chloride, magnesium, and calcium. Effect: Management of edema associated with CHF, nephrotic syndrome, and hepatic cirrhosis. I.V. furosemide may also be used as an adjunct in treatment of acute pulmonary edema. |
Mechanistic genes:COL1A1, FOS, GABRA6, IFNA1, IGF1, KDR, LTA, MMP2, NOS3, PDGFRA, PDGFRB, PTGER4, PTGS1, PTGS2, REN, SCN1B, SCNN1B, SCNN1G, TGFB1, TNFRSF1A, TNFRSF1B, VCAM1, VEGFA Metabolic genes Substrate:UGT1A1, UGT1A3, UGT1A7, UGT1A10 Transporter genes:ABCC2, ABCC3, ABCC4, SLC12A1, SLC12A3, SLC22A6, SLC22A7 Pleiotropic genes:IL6, IL10, TNF |
|
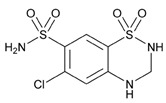
Name: HYDROCHLOROTHIAZIDE IUPAC Name: 2H-1,2,4-benzothiadiazine-7-sulfonamide, 6-chloro-3,4-dihydro-, 1,1-dioxide; 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide Molecular Formula: C7H8ClN3O4S2 Molecular Weight: 297.74 Mechanism: Inhibits sodium reabsorption in distal tubules causing increased excretion of sodium and water as well as potassium and hydrogen ions. Effect: Management of mild-to-moderate hypertension. Treatment of edema in congestive heart failure and nephrotic syndrome. |
Mechanistic genes:ACE1, ACE2, ADD1, ADRB1, ADRB2, AGT, GNB3, GRIA3, NOS3, PTGS2, REN, SCNN1G, WNK1 Metabolic genes Substrate:CYP11B2 Transporter genes:ABCC4, SLC22A6 |
|
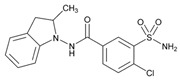
Name: INDAPAMIDE IUPAC Name: Benzamide, 3-(aminosulfonyl)-4-chloro-N-(2,3-dihydro-2-methyl-1H-indol-1-yl)-; 4-chloro-N-(2-methyl-1-indolinyl)-3-sulfamoylbenzamide Molecular Formula: C16H16ClN3O3S Molecular Weight: 365.83 Mechanism: Diuretic effect localized at proximal segment of distal tubule of nephron. Enhances sodium, chloride, and water excretion by interfering with transport of sodium ions across renal tubular epithelium. Effect: Management of hypertension, or of edema associated with CHF and nephrotic syndrome. |
Mechanistic genes:KCNE1, KCNQ1 Metabolic genes Substrate:CYP3A4, CYP3A5 |
|
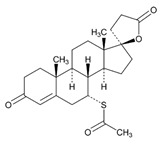
Name: SPIRONOLACTONE IUPAC Name: Pregn-4-ene-21-carboxylic acid, 7-(acetylthio)-17-hydroxy-3-oxo-, γ-lactone, (7α,17α)-; 17-hydroxy-7α-mercapto-3-oxo-17α-pregn-4-ene-21-carboxylic acid, γ-lactone acetate Molecular Formula: C24H32O4S Molecular Weight: 416.57 Mechanism: Synthetic steroid mineralocorticoid receptor antagonist (aldosterone antagonist). Exhibits magnesium- and potassium-sparing, natriuretic, diuretic, and hypotensive effects by competitively inhibiting physiologic effects of adrenocorticortical hormone aldosterone on distal renal tubules, myocardium, and vasculature. Does not generally cause potassium depletion or affect glucose metabolism or uric acid excretion. Androgen and progesterone receptor antagonist. Effect: Edema associated with excessive aldosterone excretion. Hypertension. Primary hyperaldosteronism. Hypokalemia. Cirrhosis accompanied by edema or ascites. Nephritic syndrome. Severe heart failure. |
Mechanistic genes:ACE1, AR, NR3C2, SCNN1G Metabolic genes Substrate:CYP2C8, CYP3A4, CYP3A5, CYP7A1, UGT1A1, UGT1A6 Inhibitor:CYP11B2 Transporter genes:ABCB1, ABCB11, ABCC2, ABCC3 |
|
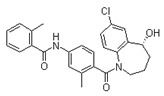
Name: TOLVAPTAN IUPAC Name: N-[4-(7-chloro-5-hydroxy-2,3,4,5-tetrahydro-1-benzazepine-1-carbonyl)-3-methylphenyl]-2-methylbenzamide; (±)-4′-[(7-chloro-2,3,4,5-tetrahydro-5-hydroxy-1H-1-benzazepin-1-yl)carbonyl]-o-tolu-m-toluidide Molecular Formula: C26H25ClN2O3 Molecular Weight: 448.94 Mechanism: Tolvaptan is a selective vasopressin V2-receptor antagonist with an affinity for the V2 receptor that is 1.8-fold that of native arginine vasopressin (AVP). Tolvaptan affinity for the V2 receptor is 29-fold greater than for the V1a receptor. Antagonism of the V2 receptor by tolvaptan promotes the excretion of free water (without loss of serum electrolytes) resulting in net fluid loss, increased urine output, decreased urine osmolality, and subsequent restoration of normal serum sodium levels. Tolvaptan metabolites have no or weak antagonist activity for human V2 receptors compared with tolvaptan. Effect: Treatment of clinically significant hypervolemic or euvolemic hyponatremia (associated with heart failure, cirrhosis or SIADH) with either a serum sodium <125 mEq/L or less marked hyponatremia that is symptomatic and resistant to fluid restriction. |
Mechanistic genes:AVPR2, PKD1, PKD2 Metabolic genes Substrate:ABCB1, CYP3A4, CYP3A5 Inhibitor:ABCB1 |
|
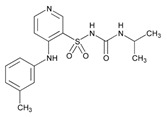
Name: TORSEMIDE IUPAC Name: 3-Pyridinesulfonamide, N-[[(1-methylethyl)amino]carbonyl]-4-[(3-methylphenyl)amino]-; 1-isopropyl-3-[(4-m-toluidino-3-pyridyl)sulfonyl]urea Molecular Formula: C16H20N4O3S Molecular Weight: 348.42 Mechanism: Inhibits reabsorption of sodium and chloride in ascending loop of Henle and distal renal tubule, interfering with chloride-binding cotransport system, thus causing increased excretion of water, sodium, chloride, magnesium, and calcium. Effect: Management of edema associated with congestive heart failure and hepatic or renal disease. Used alone or in combination with antihypertensives in treatment of hypertension. I.V. form indicated when rapid onset is desired. |
Mechanistic genes:ADD1, SCNN1G Metabolic genes Substrate:CYP2C8, CYP2C9, CYP11B2 Inhibitor:CYP2C19 Transporter genes:SLC12A1, SLC12A3, SLCO1B1 |
|
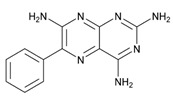
Name: TRIAMTERENE IUPAC Name: 2,4,7-Pteridinetriamine, 6-phenyl-; (2) 2,4,7-triamino-6-phenylpteridine Molecular Formula: C12H11N7 Molecular Weight: 253.26 Mechanism: Interferes with potassium/sodium exchange (active transport) in distal tubule, cortical collecting tubule and collecting duct by inhibiting sodium, potassium-ATPase. Decreases calcium excretion. Increases magnesium loss. Effect: Alone or in combination with other diuretics in treatment of edema and hypertension. Decreases potassium excretion caused by kaliuretic diuretics. |
Mechanistic genes:SCNN1A, SCNN1B, SCNN1D, SCNN1G Metabolic genes Substrate:CYP1A2, CYP2A6, CYP2D6, CYP3A4, CYP3A5 Inducer:CYP1A2 |
ABCA1: ATP-binding cassette, subfamily A, member 1; ABCB1: ATP-binding cassette, subfamily B, member 1; ABCB11: ATP-binding cassette, subfamily B, member 11; ABCC1: ATP-binding cassette, subfamily C, member 1; ABCC2: ATP-binding cassette, subfamily C, member 2; ABCC3: ATP-binding cassette, subfamily C, member 3; ABCC4: ATP-binding cassette, subfamily C, member 4; ABCC5: ATP-binding cassette, subfamily C, member 5; ABCC6: ATP-binding cassette, subfamily C, member 6; ABCC8: ATP-binding cassette, subfamily C, member 8; ABCC9: ATP-binding cassette, subfamily C, member 9; ABCG2: ATP-binding cassette, subfamily G, member 2; ABL1: ABL protooncogene 1, non-receptor tyrosine kinase; ACACA: Acetyl-CoA carboxylase-alpha; ACE1: Angiotensin I-converting enzyme; ACE2: Angiotensin I-converting enzyme 2; ACOX1: Acyl-CoA oxidase 1, palmitoyl; ACSL1: Acyl-CoA synthetase long chain family, member 1; ACSM1: Acyl-CoA synthetase medium chain family member 1; ACVRL1: Activin A receptor, type II-like 1; ADA: Adenosine deaminase; ADAMTS4: A disintegrin-like and metalloproteinase with thrombospondin type 1 motif, 4; ADD1: Adducin 1; ADIPOQ: Adipocyte-, C1q-, and collagen domain-containing; ADORA1: Adenosine A1 receptor; ADORA2A: Adenosine A2A receptor; ADORA2B: Adenosine A2b receptor; ADRA1A: Alpha-1A-adrenergic receptor; ADRA1B: Alpha-1B-adrenergic receptor; ADRA1D: Alpha-1D-adrenergic receptor; ADRA2A: Alpha-2A-adrenergic receptor; ADRA2B: Alpha-2B-adrenergic receptor; ADRA2C: Alpha-2C-adrenergic receptor; ADRB1: Beta-1-adrenergic receptor; ADRB2: Beta-2-adrenergic receptor; ADRB3: Beta-3-adrenergic receptor; AGPAT2: 1-Acylglycerol-3-phosphate o-acyltransferase 2; AGT: Angiotensinogen; AGTR1: Angiotensin II receptor, type 1; AGTR2: Angiotensin II receptor, type 2; AHR: Aryl hydrocarbon receptor; AKR1C1: Aldo-keto reductase family 1, member 1; AKR1C4: Aldo-keto reductase family 1, member C4; ALB: Albumin; ALDH2: Aldehyde dehydrogenase 2 family; ALDH3A1: Aldehyde dehydrogenase, family 3, subfamily A, member 1; ALOX5: Arachidonate 5-lipoxygenase; ANXA1: Annexin A1; ANXA2: Annexin A2; ANXA5: Annexin A5; APC: APC regulator of wnt signaling pathway; APOA1: Apolipoprotein A-I; APOA5: Apolipoprotein A-V; APOB: Apolipoprotein B; APOC3: Apolipoprotein C-III; APOE: Apolipoprotein E; APP: Amyloid beta A4 precursor protein; AR: Androgen receptor; ATP1A1: ATPase, Na+/K+ transporting, alpha-1 polypeptide; AVPR1A: Arginine vasopressin receptor 1A; AVPR2: Arginine vasopressin receptor 2; BCAR1: BCAR1 scaffold protein, CAS family member; BDKRB2: Bradykinin receptor B2; BMPR2: Bone morphogenetic protein receptor, type II; CA7: Carbonic anhydrase VII; CA12: Carbonic anhydrase XII; CA13: Carbonic anhydrase XIII; CACNA1C: Calcium channel, voltage-dependent, L type, alpha-1C subunit; CACNA1D: Calcium channel, voltage-dependent, L type, alpha-1D subunit; CACNA2D1: Calcium channel, voltage-dependent, alpha-2/delta subunit 1; CACNB2: Calcium channel, voltage-dependent, beta-2 subunit; CACNG1: Calcium channel, voltage-dependent, gamma-1 subunit; CALR: Calreticulin; CALU: Calumenin; cAMP: Cyclic adenosine monophosphate; CANX: Calnexin; CASP1: Caspase 1, apoptosis-related cysteine protease; CASP3: Caspase 3, apoptosis-related cysteine protease; CAT: Catalase; CBR1: Carbonyl reductase 1; CBS: Cystathionine beta-synthase; CCNA2: Cyclin A2; CCND1: Cyclin D1; CDK2: Cyclin-dependent kinase 2; CDK4: Cyclin-dependent kinase 4; CEL: Carboxyl-ester lipase; CES1: Carboxylesterase 1; CES2: Carboxylesterase 2; CETP: Cholesteryl ester transfer protein, plasma; CFH: Complement factor H; CFTR: Cystic fibrosis transmembrane conductance regulator; CHAT: Choline acetyltransferase; CHRM2: Cholinergic receptor, muscarinic, 2; CHRNA1: Cholinergic receptor, nicotinic, alpha polypeptide 1; CHRNA2: Cholinergic receptor, neuronal nicotinic, alpha polypeptide 2; CHRNA4: Cholinergic receptor, neuronal nicotinic, alpha polypeptide 4; CHST3: Carbohydrate sulfotransferase 3; CLEC3B: Tetranectin; CMBL: Carboxymethylenebutenolidase-like protein; CNP: Cyclic nucleotide phosphodiesterase; CNR1: Cannabinoid receptor 1; COL1A1: Collagen, type I, alpha-1; COMT: Catechol-O-methyltransferase; COX-1: Cycloxygenase-1; CPA1: Carboxypeptidase A1; CREB1: cAMP response element-binding protein 1; CRP: C-reactive protein; CRYZ: Crystallin, zeta; CXCL12: Chemokine, CXC motif, ligand 12; CYP1A1: Cytochrome P450, subfamily I, polypeptide 1; CYP1A2: Cytochrome P450, subfamily I, polypeptide 2; CYP1B1: Cytochrome P450, subfamily I, polypeptide 1; CYP2A6: Cytochrome P450, subfamily IIA, polypeptide 6; CYP2B6: Cytochrome P450, subfamily IIB, polypeptide 6; CYP2C8: Cytochrome P450, subfamily IIC, polypeptide 8; CYP2C9: Cytochrome P450, subfamily IIC, polypeptide 9; CYP2C18: Cytochrome P450, subfamily IIC, polypeptide 18; CYP2C19: Cytochrome P450, subfamily IIC, polypeptide 19; CYP2D6: Cytochrome P450, subfamily IID, polypeptide 6; CYP2E1: Cytochrome P450, subfamily IIE; CYP2J2: Cytochrome P450, subfamily IIJ, polypeptide 2; CYP3A4: Cytochrome P450, subfamily IIIA, polypeptide 4; CYP3A5: Cytochrome P450, subfamily IIIA, polypeptide 5; CYP3A7: Cytochrome P450, subfamily IIIA, polypeptide 7; CYP4F2: Cytochrome P450, family 4, subfamily F, polypeptide 2; CYP7A1: Cytochrome P450, subfamily VIIA, polypeptide 1; CYP11A1: Cytochrome P450, subfamily XIA, polypeptide 1; CYP11B1: Cytochrome P450, Subfamily XIB, polypeptide 1; CYP11B2: Cytochrome P450, subfamily XIB, polypeptide 2; CYP17A1: Cytochrome P450, family 17, subfamily A, polypeptide 1; CYP19A1: Cytochrome P450, family 19, subfamily A, polypeptide 1; CYP27A1: Cytochrome P450, subfamily XXVIIA, polypeptide 1; DDC: Dopa decarboxylase; DIO2: Deiodinase iodothyronine, type II; DRD1: Dopamine receptor D1; DRD2: Dopamine receptor D2; EDN1: Endothelin 1; EDNRA: Endothelin receptor, type A; EDNRB: Endothelin receptor, type B; EGFR: Epidermal growth factor receptor; EPHX1: Epoxide hydrolase 1; ERAP1: Endoplasmic reticulum aminopeptidase 1; ERBB2: ERB-B2 receptor tyrosine kinase 2; ERBB4: ERB-B2 receptor tyrosine kinase 4; ESR1: Estrogen receptor 1; F2: Coagulation factor II; F2R: Coagulation factor II receptor; F5: Coagulation factor V; F7: Coagulation factor VII; F9: Coagulation factor IX; F10: Coagulation factor X; F13A1: Factor XIII, A1 subunit; FABP1: Fatty acid-binding protein 1; FABP2: Fatty acid-binding protein 2; FCGR2A: Fc fragment of IgG receptor IIa; FCGR2B: Fc fragment of IgG receptor IIb; FCGR3A: Fc fragment of IgG receptor IIIa; FGA: Fibrinogen, A alpha polypeptide; FGB: Fibrinogen, B beta polypeptide; FGF1: Fibroblast growth factor 1; FGF2: Fibroblast growth factor 2; FGF4: Fibroblast growth factor 4; FGF19: Fibroblast growth factor 19; FGFR1: Fibroblast growth factor receptor 1; FGFR2: Fibroblast growth factor receptor 2; FGFR4: Fibroblast growth factor receptor 4; FKBP5: FK506-binding protein 5; FOS: FOS protooncogene, AP1 transcription factor subunit; G6PD: Glucose-6-phosphate dehydrogenase; GABRA6: Gamma-aminobutyric acid receptor, alpha-6; GGCX: Gamma-glutamyl carboxylase; GLUL: Glutamate-ammonia ligase; GLYAT: Glycine-N-acyltransferase; GNAS: GNAS complex locus; GNB3: Guanine nucleotide-binding protein, beta-3; GP1BA: Glycoprotein Ib platelet subunit Alpha; GP5: Glycoprotein V, platelet; GP6: Glycoprotein VI platelet; GRIA3: Glutamate receptor, ionotropic, AMPA 3; GRK5: G protein-coupled receptor kinase 5; GSTA1: Glutathione S-transferase, alpha-1; GSTK1: Glutathione S-transferase, kappa-1; GSTM1: Glutathione S-transferase, MU-1; GSTP1: Glutathione S-transferase, PI; GSTs: Glutathione S-transferase family; GSTT1: Glutathione S-transferase, theta-1; HBB: Hemoglobin-beta locus; HCII: Heparin cofactor II; HCN1: Hyperpolarization-activated cyclic nucleotide-gated potassium channel 1; HCN4: Hyperpolarization-activated cyclic nucleotide-gated potassium channel 4; HGF: Hepatocyte growth factor; HFE: Homeostatic iron regulator; HIF1A: Hypoxia-inducible factor 1, alpha subunit; HLA-A: Major histocompatibility complex, class I, A; HLA-B: Major histocompatibility complex, class I, B; HMGCR: 3-Hydroxy-3-methylglutaryl-CoA reductase; HNF4A: Hepatocyte nuclear factor 4-alpha; HPSE: Heparanase; HRH1: Histamine receptor H1; HRH2: Histamine receptor H2; HSPA5: Heat-shock 70-kd protein 5; HTR1A: 5-hydroxytryptamine receptor 1A; HTR1B: 5-hydroxytryptamine receptor 1B; HTR3B: 5-Hydroxytryptamine receptor 3B; HTR3C: 5-Hydroxytryptamine receptor 3C; ICAM1: Intercellular adhesion molecule 1; IFNA1: Interferon, alpha-1; IFNG: Interferon, gamma; IGF1: Insulin-like growth factor I; IKBKB: Inhibitor of nuclear factor kappa-B kinase, subunit beta; IL1B: Interleukin 1-beta; IL1RN: Interleukin 1 receptor antagonist; IL2: Interleukin 2; IL4: Interleukin 4; IL6: Interleukin 6; IL8RB: Interleukin 8 receptor, beta; IL10: Interleukin 10; IL12B: Interleukin 12B; ITGA2B: Integrin, alpha-2b; ITGA4: Integrin, alpha-4; ITGB3: Integrin, beta-3; KCNA5: Potassium channel, voltage-gated, shaker-related subfamily, member 5; KCNE1: Potassium channel, voltage-gated, isk-related subfamily, member 1; KCNE2: Potassium channel, voltage-gated, isk-related subfamily, member 2; KCNH2: Potassium channel, voltage-gated, subfamily H, member 2; KCNJ1: Potassium channel, inwardly rectifying, subfamily J, member 1; KCNJ5: Potassium channel, inwardly rectifying, subfamily J, member 5; KCNJ11: Potassium channel, inwardly rectifying, subfamily J, member 11; KCNMB1: Potassium channel, calcium-activated, large conductance, subfamily M, beta member 1; KCNQ1: Potassium channel, voltage-gated, KQT-like subfamily, member 1; KDR: Kinase insert domain receptor; KRT8: Keratin 8, type II; LDLR: Low-density lipoprotein receptor; LEP: Leptin; LIPA: Lipase A, lysosomal acid; LIPC: Lipase, hepatic; LMW: Low molecular weight; LPL: Lipoprotein lipase; LRP1: Low-density lipoprotein receptor-related protein 1; LRP2: Low-density lipoprotein receptor-related protein 2; LTA: Lymphotoxin-alpha; MAOA: Monoamine oxidase A; MAOB: Monoamine oxidase B; MAP2K4: Mitogen-activated protein kinase kinase 4; MAPT: Microtubule-associated protein tau; MGMT: Methylguanine-DNA methyltransferase; MMP2: Matrix metalloproteinase 2; MMP3: Matrix metalloproteinase 3; MMP12: Matrix metalloproteinase 12; MPO: Myeloperoxidase; MTHFR: 5,10-Methylenetetrahydrofolate reductase; MT-ND4: Complex I, subunit ND4; MTR: 5-Methyltetrahydrofolate-homocysteine S-methyltransferase; MTTP: Microsomal triglyceride transfer protein; MYC: MYC protooncogene, bHLH transcription factor; NAT2: N-acetyltransferase 2; NFKB1: Nuclear factor kappa-B, subunit 1; NFKBIA: Nuclear factor kappa-B inhibitor, alpha; NID1: Nidogen 1; NNMT: Nicotinamide N-methyltransferase; NOS1AP: Nitric oxide synthase 1 (neuronal) adaptor protein; NOS2: Nitric oxide synthase 2; NOS3: Nitric oxide synthase 3; NOX1: NADPH oxidase 1; NPC1: NPC intracellular cholesterol transporter 1; NPC1L1: NPC1-like intracellular cholesterol transporter 1; NPPA: Natriuretic peptide precursor A; NPR1: Natriuretic peptide receptor a/guanylate cyclase A; NPR2: Natriuretic peptide receptor 2; NPR3: Natriuretic peptide receptor 3; NPY: Neuropeptide Y; NQO1: NAD(P)H dehydrogenase, quinone 1; NR1I2: Nuclear receptor subfamily 1, group I, member 2; NR1I3: Nuclear receptor subfamily 1, group I, member 3; NR3C1: Nuclear receptor subfamily 3, group C, member 1; NR3C2: Nuclear receptor subfamily 3, group C, member 2; ORM1: Orosomucoid 1; P2RY1: Purinergic receptor P2Y, g protein-coupled, 1; P2RY12: Purinergic receptor P2RY12; PAF-1: Plasminogen activator inhibitor-1; PAR-1: Protease-activated receptor 1; PARK2: Parkin; PCNA: Proliferating cell nuclear antigen; PDE1C: Phosphodiesterase 1C; PDE3A: Phosphodiesterase 3A; PDE4A: Phosphodiesterase 4A; PDE4B: Phosphodiesterase 4B; PDE4C: Phosphodiesterase 4C; PDE4D: Phosphodiesterase 4D; PDE5A: Phosphodiesterase 5A; PDE10A: Phosphodiesterase 10A; PDGFR: Platelet-derived growth factor receptor; PDGFRA: Platelet-derived growth factor receptor, alpha; PDGFRB: Platelet-derived growth factor receptor, beta; PF4: Platelet factor 4; PGD2: Prostaglandin D2; PGE2: Prostaglandin E2; PGI2: Prostacyclin; PKA: Protein kinase A; PKD1: Polycystin 1; PKD2: Polycystin 2; PLA2G4A: Phospholipase A2, group IVA; PLA2s: Phospholipase A2 family; PLAT: Plasminogen activator, tissue; PLAU: Plasminogen activator, urinary; PLAUR: Plasminogen activator receptor, urokinase-type; PLG: Plasminogen; POMC: Proopiomelanocortin; PON1: Paraoxonase 1; POR: Cytochrome P450 oxidoreductase; PPARA: Peroxisome proliferator-activated receptor-alpha; PPARD: Peroxisome proliferator-activated receptor-delta; PPARG: Peroxisome proliferator-activated receptor-gamma; PPARGC1A: Peroxisome proliferator-activated receptor-gamma, coactivator 1, alpha; PRKRA: Protein kinase, interferon-inducible double-stranded MA-dependent activator; PRNP: Prion protein; PROC: Protein C; PROCR: Protein c receptor; PROS1: Protein S; PSEN1: Presenilin 1; PTGDR2: Prostaglandin D2 receptor 2; PTGER1: Prostaglandin E receptor 1; PTGER2: Prostaglandin E receptor 2, EP2 subtype; PTGER3: Prostaglandin E receptor 3, EP3 subtype; PTGER4: Prostaglandin E receptor 4, EP4 subtype; PTGES: Prostaglandin E synthase; PTGIR: Prostaglandin I2 receptor; PTGIS1: Prostaglandin-endoperoxide synthase 1; PTGIS: Prostaglandin I2 synthase; PTGIS2: Prostaglandin-endoperoxide synthase 2; PTGS1: Prostaglandin-endoperoxide synthase 1; PTGS2: Prostaglandin-endoperoxide synthase 2; RAP1B: Ras family small GTP binding protein RAP1B; RB1: RB transcriptional corepressor 1; RCAN1: Regulator of calcineurin 1; REN: Renin; RET: RET protooncogene; RPS6KA3: Ribosomal protein S6 kinase A3; RYR1: Ryanodine receptor 1; SCARB1: Scavenger receptor class B, member 1; SCN1B: Sodium voltage-gated channel, beta subunit 1; SCN5A: Sodium voltage-gated channel, alpha subunit 5; SCN10A: Sodium voltage-gated channel, alpha subunit 10; SCNN1A: Sodium channel, epithelial 1, alpha subunit; SCNN1B: Sodium channel, epithelial 1, beta subunit; SCNN1D: Sodium channel, epithelial 1, delta subunit; SCNN1G: Sodium channel, epithelial 1, gamma subunit; SELP: Selectin P; SERPINA5: Serpin peptidase inhibitor, clade A, member 5; SERPINA6: Serpin peptidase inhibitor, clade A, member 6; SERPINA7: Serpin peptidase inhibitor, clade A, member 7; SERPINB2: Serpin peptidase inhibitor, clade B (ovalbumin), member 2; SERPINB6: Serpin peptidase inhibitor, clade B (ovalbumin), member 6; SERPINC1: Serpin peptidase inhibitor, clade C (antithrombin), member 1; SERPIND1: Heparin cofactor II; SERPINE1: Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1; SHBG: Sex hormone-binding globulin; SLC5A5: Solute carrier family 5 (sodium iodide symporter), member 5; SLC5A8: Solute carrier family 5 (iodide transporter), member 8; SLC6A2: Solute carrier family 6 (neurotransmitter transporter, noradrenaline), member 2; SLC6A3: Solute carrier family 6 (neurotransmitter transporter, noradrenaline), member 3; SLC10A1: Solute carrier family 10 (sodium/bile acid cotransporter family), member 1; SLC12A1: Solute carrier family 12 (sodium/potassium/chloride transporter), member 1; SLC12A3: Solute carrier family 12 (sodium/chloride transporter), member 3; SLC14A2: Solute carrier family 14 (urea transporter), member 2; SLC15A1: Solute carrier family 15 (oligopeptide transporter), member 1; SLC18A2: Solute carrier family 18 (vesicular monoamine), member 2; SLC19A1: Solute carrier family 19 (folate transporter), member 1; SLC22A1: Solute carrier family 22 (organic cation transporter), member 1; SLC22A2: Solute carrier family 22 (organic cation transporter), member 2; SLC22A6: Solute carrier family 22 (organic anion transporter), member 6; SLC22A7: Solute carrier family 22 (organic anion transporter), member 7; SLC22A8: Solute carrier family 22 (organic anion transporter), member 8; SLC22A16: Solute carrier family 22 (organic cation transporter), member 16; SLCO1A2: Solute carrier organic anion transporter family, member 1A2; SLCO1B1: Solute carrier organic anion transporter family, member 1B1; SLCO1B2: Solute carrier organic anion transporter family, member 1B2; SLCO1B3: Solute carrier organic anion transporter family, member 1B3; SLCO2A1: Solute carrier organic anion transporter family, member 2A1; SLCO2B1: Solute carrier organic anion transporter family, member 2B1; SLCO3A1: Solute carrier organic anion transporter family, member 3A1; SNCA: Synuclein, alpha; SOD2: Superoxide dismutase 2; ST14: Suppression of tumorigenicity 14; STAT3: Signal transducer and activator of transcription 3; SULT1A1: Sulfotransferase family 1A, cytosolic, phenol-preferring, member 1; SULT1A3: Sulfotransferase family 1A, cytosolic, phenol-preferring, member 3; SULT1E1: Sulfotransferase family 1E, estrogen-preferring, member 1; TBX21: T-box transcription factor 21; TBXA2R: Thromboxane A2 receptor; TBXAS1: Thromboxane A synthase 1; TCF20: Transcription factor 20; TFPI: Tissue factor pathway inhibitor; TGFB1: Transforming growth factor, beta-1; TGFBR1: Transforming growth factor-beta receptor, type I; THBD: Thrombomodulin; TNF: Tumor necrosis factor; TNFAIP6: Tumor necrosis factor-alpha-induced protein 6; TNFRSF1A: Tumor necrosis factor Receptor superfamily, member 1A; TNFRSF1B: Tumor necrosis factor receptor subfamily, member 1B; TP53: Tumor protein p53; TPMT: Thiopurine s-methyltransferase; TRAP: Thrombin receptor agonist peptide; TRPV1: Transient receptor potential cation channel, subfamily v, member 1; UCP2: Uncoupling protein 2; UGT1A1: UPD-glycosyltransferase 1 family, polypeptide A1; UGT1A3: UPD-glycosyltransferase 1 family, polypeptide A3; UGT1A4: UDP-glycosyltransferase 1 family, polypeptide A4; UGT1A5: UDP-glycosyltransferase 1 family, polypeptide A5; UGT1A6: UDP-glycosyltransferase 1 family, polypeptide A6; UGT1A7: UDP-glycosyltransferase 1 family, polypeptide A7; UGT1A9: UDP-glycosyltransferase 1 family, polypeptide A9; UGT1A10: UDP-glycosyltransferase 1 family, polypeptide A10; UGT2B4: UDP glucuronosyltransferase family 2, member B4; UGT2B7: Uridine diphosphate glycosyltransferase 2 family, member B7; UGT2B15: Uridine diphosphate glycosyltransferase 2 family, member B15; UGT2B17: Uridine diphosphate glycosyltransferase 2 family, member B17; UGTs: UDP glucuronosyltransferase family; USP5: Ubiquitin-specific protease 5; VCAM1: Vascular cell adhesion molecule 1; VEGFA: Vascular endothelial growth factor A; VKORC1: Vitamin K epoxide reductase complex subunit 1; VTN: Vitronectin; VWF: von Willebrand factor; WNK1: Protein kinase, lysine-deficient 1; XDH: Xanthine dehydrogenase.
