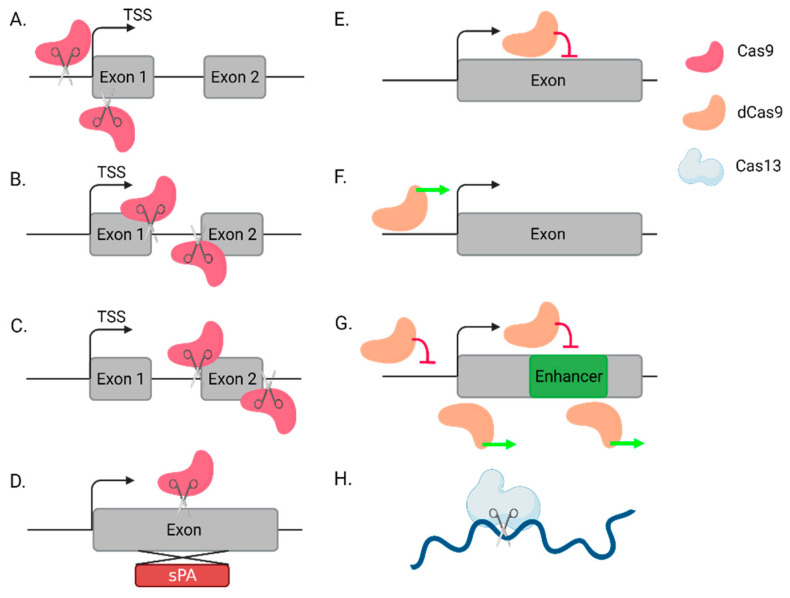
Figure 1.
A schematic illustration of CRISPR-based approaches to study lncRNAs. (A) Transcription start site (TSS) deletion. (B) Mutations of splice sites. (C) Removal of an exon or a large genomic fragment. (D) Knock-in: insertion of a synthetic polyadenylation (spA) signal. (E) Knockdown by CRISPRi. (F) Gene overexpression by CRISPRa. (G) Tiling a genomic locus using CRISPR-ko/CRISPRi/CRISPRa to identify enhancers, which in many cases can overlap or affect lncRNAs. (H) Targeting an RNA transcript by Cas13. (Created with BioRender.com (accessed on 12 December 2021)).