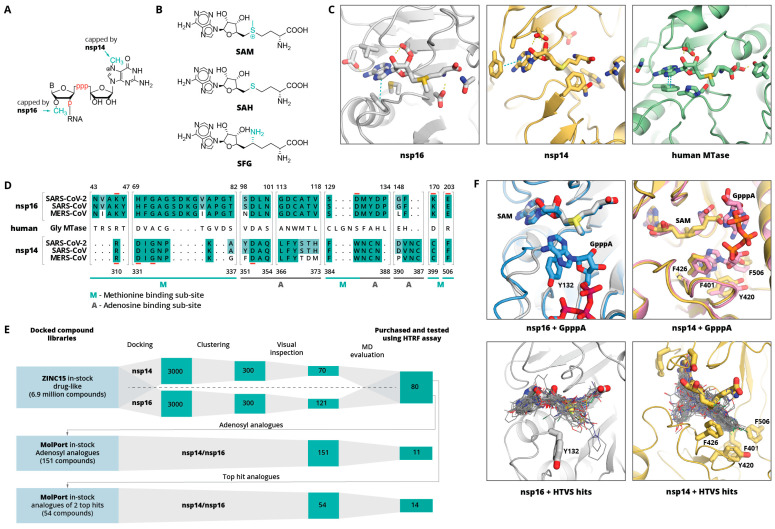
Figure 1.
(A) Schematic representation of nsp14 and nsp16 mRNA capping sites. B indicates base, p indicates phosphate group. (B) Molecular structures of methyl group donor S-adenosylmethionine (SAM), the reaction product S-adenosylhomocysteine (SAH), and the SAH analogue sinefungin (SFG). (C) Binding sites of SARS-CoV-2 nps16 (PDB ID: 6W4H), SARS-CoV nsp14 (PDB ID: 5C8T), and human glycine N-methyltransferase (PBD ID: 1R74 [38]). Bound SAM and key binding site residues are shown as sticks. Yellow dashed lines indicate hydrogen bonds, cyan—aromatic stacking. (D) Sequence alignment of nsp14 and nsp16 binding sites of SARS-CoV-2, SARS-CoV, and MERS CoV. Shading indicates the conservation of residues with fully conserved positions shaded in darker green. An aligned sequence of the human glycine N-methyltransferase is given in the middle. Residue numbers on top refer to the SARS-CoV-2 nsp16 and on the bottom refer to SARS-CoV nsp14. Active site residues are underlined in red. The adenine and methionine fragment binding sub-sites are indicated with A or M, respectively. (E) Schematic workflow of the computational inhibitor discovery approach used. Number of compounds retained at each stage is shown in green rectangles. The workflow was carried out for both targets separately, and the identified virtual screening hits were tested experimentally against both enzymes. (F) Structural alignment of GpppA-bound and unbound nsp14 and nsp16 complexes with SAM (top) and 50 top-scoring HTVS hits (bottom). SAM, GpppA, and hydrophobic amino acids interacting with RNA base are shown as sticks, HTVS hits are shown as lines.