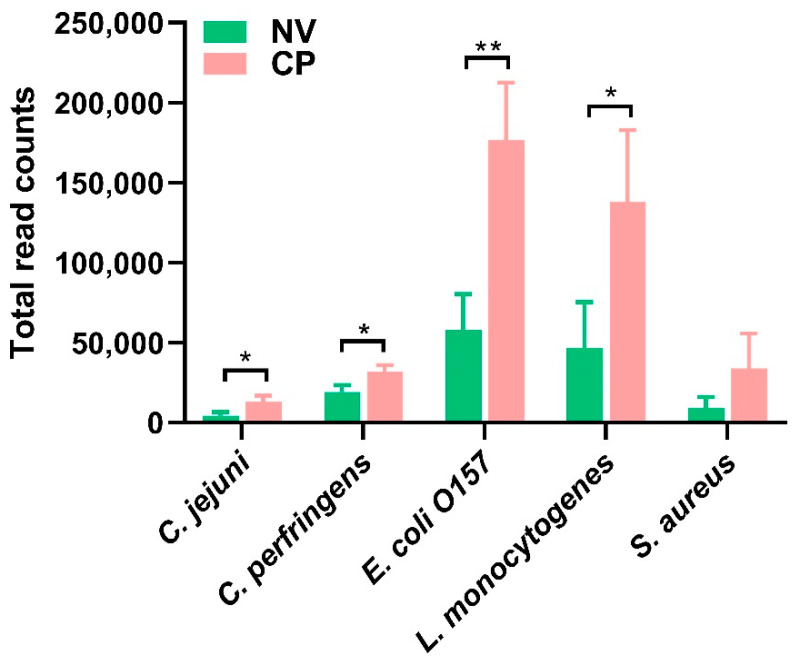
Figure 4.
The relative ratios (*103) of transcriptional read counts of five major foodborne pathogens to total counts in small intestines at Day 5 post C. perfringens infections (twice a day for five days by oral gavage). The RNA was extracted from contents, and mRNA was sequenced by RNA-seq. Statistical analysis for each bacterium in C. perfringens (CP) infected group was carried out using unpaired T-test. * means p < 0.05 and ** mean p < 0.01 when compared to the sham control (NV) group. These bacterial references used were Listeria monocytogenes (Gene Bank accession number CP019624.1), Campylobacter jejuni (AL111168.1), Staphylococcus aureus (BX571856.1), and Escherichia coli O157 (CP043539.1), and C. perfringens (BA000016.3).