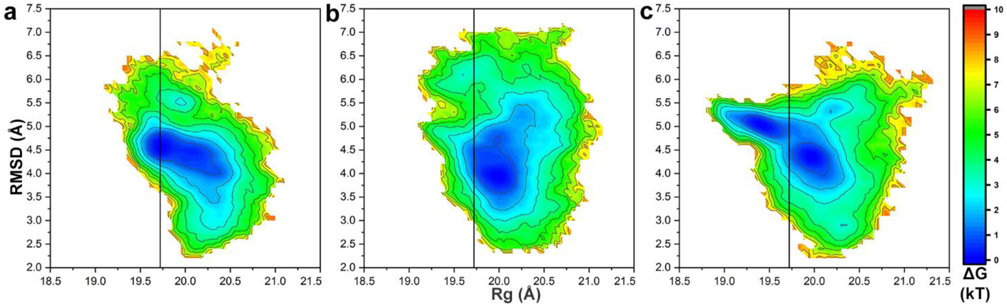
Fig. 2.
Conformational landscape sampled by the capsid protein CP∗ 42-231. Free energy surface as function of parameters RMSD (CP∗ 42-231 Cα atoms) vs. Rg (CP∗ 42-231 Cα atoms) from the simulation at (a) pH 3, (b) pH 5, (c) pH 7. RMSD was calculated by comparing conformations generated from 1 μsec of simulation to the reference CP in the assembled capsid (PDB entry 3R0R). Vertical black lines signify Rg value of PDB entry 3R0R.