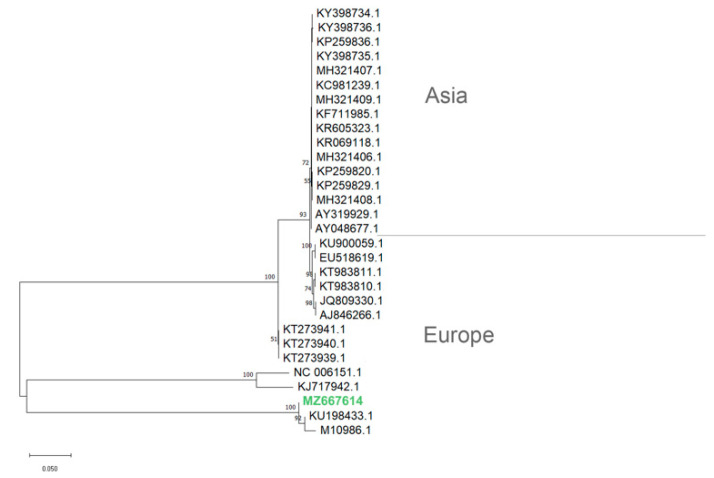
Figure 5.
Phylogenetic tree (NJ) depicting relationships among the PrV isolates based on glycoprotein G genes. Nucleotide sequences were aligned using the ClustalW algorithm, and the evolutionary history was inferred using the Neighbor Joining (NJ) method in MEGA11 (Tamura et al., 2021). Bootstrap values (1000 replicates) are indicated at the nodes. GenBank accessions from PrV strains are as follow (accession/strain): KU198433.1/ADV32751/Italy2014; NC_006151.1/, KJ717942.1/ and AJ846266.1/Kaplan; KT273939.1/SuHV1/WB6/NIVS2014; KT273940.1/SuHV1/WB7/NIVS2014; KT273941.1/SuHV1/WB8/NIVS2014; JQ809330.1/DUL34Pass; KT983810.1/Hercules; KT983811.1/Kolchis; EU518619.1/ and KU900059.1/NiA3; MH321406.1/HN-HH, MH321407.1/HN-NY, MH321408.1/HN-NX, MH321409.1/HN-ZZ, KP259820.1/YY, KP259829.1/LY, KP259836.1/GA, KR069118.1/isolate BJ, KR605323.1/AH02LA, KF711985.1/Xiang A, KC981239.1/BJ/YT, KY398734.1/isolate SCT17-11, KY398735.1/isolate HuNF83-14, KY398736.1/isolate SDZD-16, M10986.1, AY319929.1/Ea, and AY048677.1.