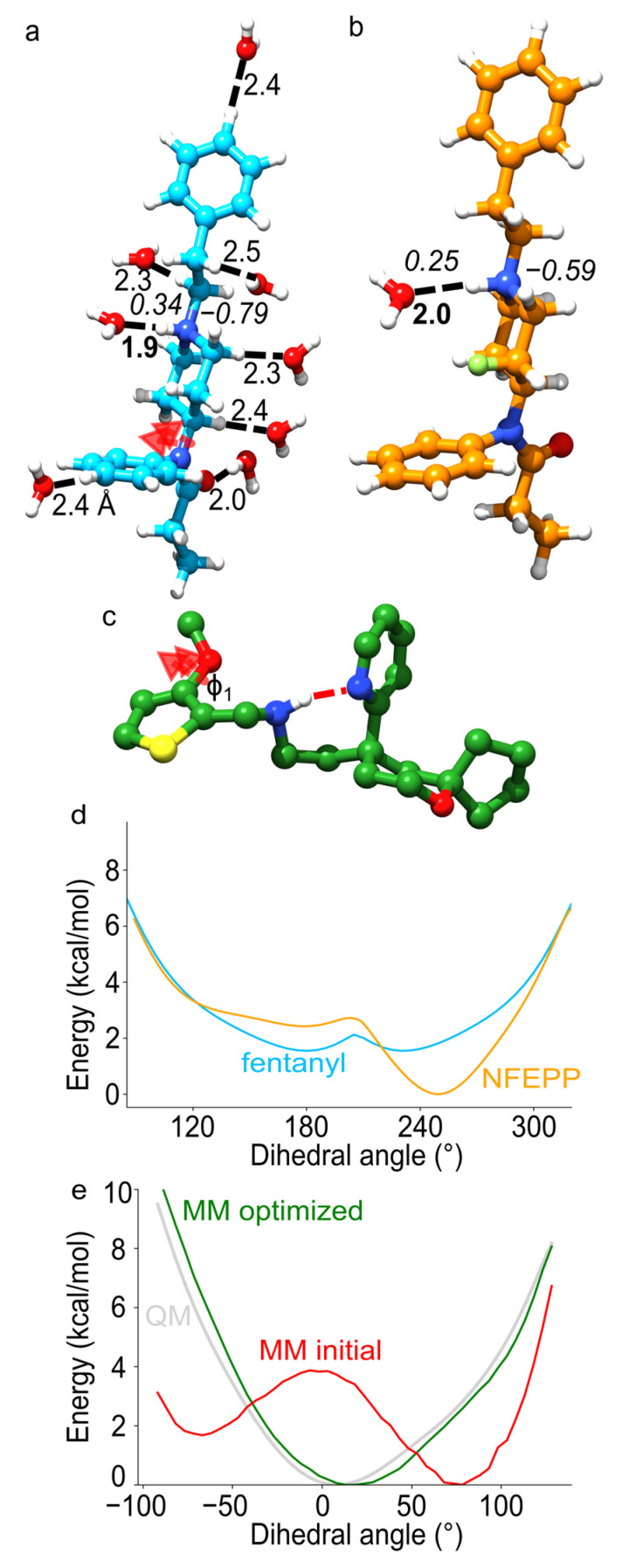
Figure 7.
Force-field parameterization of fentanyl, NFEPP and oliceridine. (a) Illustration of QM interaction distances (regular fonts) and atomic partial charges (italic fonts) for selected sites of fentanyl. Note that the protonated amine can H-bond to water. The arrow indicates the bond for which we illustrate in panel d the torsional energy profile. (b) H-bond distance and partial charge values for the charged amine group in NFEPP. (c) Coordinate snapshot from MM MD of oliceridine [88] illustrating an internal H-bond (red dotted line) sampled between the piperidine and secondary amine nitrogen atoms. Dihedral angle Φ1 is flexible and required force-field parametrization. (d) Illustration of the potential energy scans for a bond twist parametrized for fentanyl and NFEPP. Potential energies, taken relative to the energy of the equilibrium geometry, were computed with MM parameters derived in Ref. [87]. (e) Potential energy scans computed for angle Φ1. The initial MM profile is colored red, QM target profile, gray, and MM optimized, green. Panel e is based on data published in Ref [88]. Molecular graphics in panel a, b and c were created with UCSF Chimera 1.14 [23], whereas the text, arrows and dotted lines were added with Inkscape 1.0. Plots in d and e were generated using Python’s matplotlib package [89].