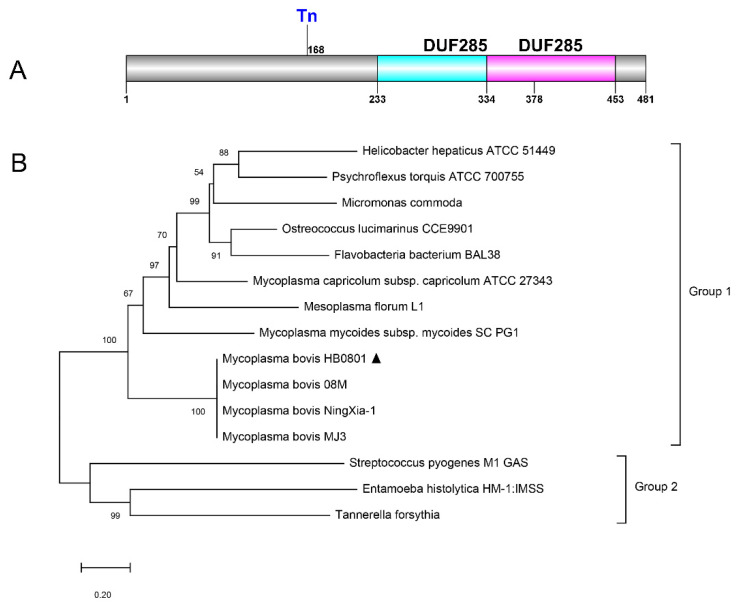
Figure 1.
Overall gene organization of Mbov_0145 and its phylogenetic analysis. (A) Schematic representation of the domain structure of MbovP0145 via the NCBI database. (B) Phylogeny of MbovP0145 and orthologs from other related bacteria. The trees were constructed using the neighbor-joining method with nodal support assessed by 1000 bootstrap replicates. Bacteria from which complete SPase I sequences were taken and their respective GenBank or NCBI reference sequence accession numbers are as follows: M. bovis HB0801 (AFM51518.1), M. bovis 08M (AQU85457.1), M. bovis NingXia-1 (ATQ40684.1), M. bovis MJ3 (AXJ69624.1), Mycoplasma mycoides subsp. mycoides SC PG1 (WP_011167164.1), Mycoplasma capricolum subsp. capricolum ATCC 27343 (WP_011387196.1), Mesoplasma florum L1 (WP_011183344.1), Ostreococcus lucimarinus CCE9901 (XP_001422294.1), Helicobacter hepaticus ATCC 51449 (WP_011114893.1), Psychroflexus torquis ATCC 700755 (ZP_01255287.1), Micromonas commode (ACO63939.1), Flavobacteria bacterium BAL38 (ZP_01734433.1), Streptococcus pyogenes M1 GAS (AAK33772.1), Entamoeba histolytica HM-1:IMSS (XP_653526.1), and Tannerella forsythia (WP_157755308.1). M. bovis HB0801 is indicated by the triangle marker “▲”.