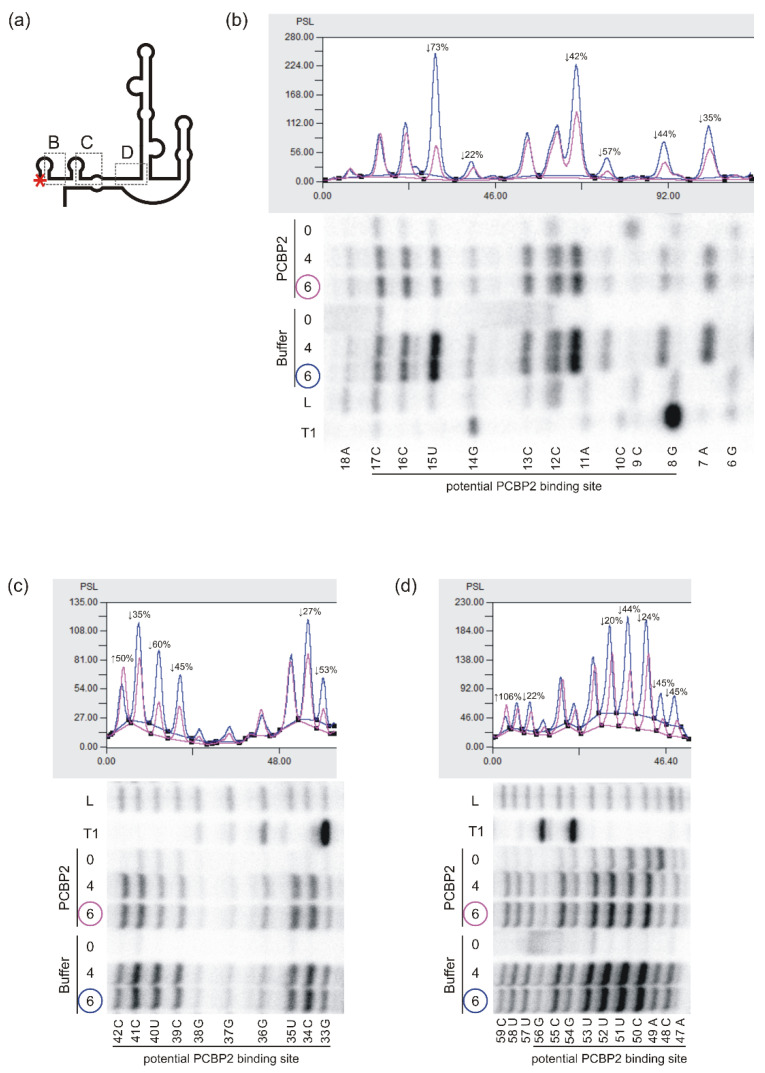
Figure 4.
Structural probing of the P1-Δ40p53 RNA in vitro in the presence of PCBP2 by Pb2+-induced cleavage method. (a) Schematic representation of the structure of the P1-Δ40p53 RNA. The red asterisk denotes the 5′-[32P]-end-labelling of RNA. The analysed regions of RNA are marked with dotted-line boxes named B, C and D. (b–d) Bottom panels: the autoradiograms show the products of Pb2+-ions induced cleavage reactions analysed on 12% polyacrylamide gels in denaturing conditions. The reactions were carried out at 37 °C with 4 and 6 mM Pb2+ ions for 3 min in the presence of PCBP2 or the buffer. Lanes: L, formamide ladder; T1, limited digestion by RNase T1 in denaturing conditions. Nucleotide residues are marked on the left side of each autoradiogram. Black lines along the gels indicate potential PCBP2 binding sites. Upper panels: normalised reactivities of Pb2+-induced cleavage (6 mM Pb2+ concentration) as a function of nucleotide position.