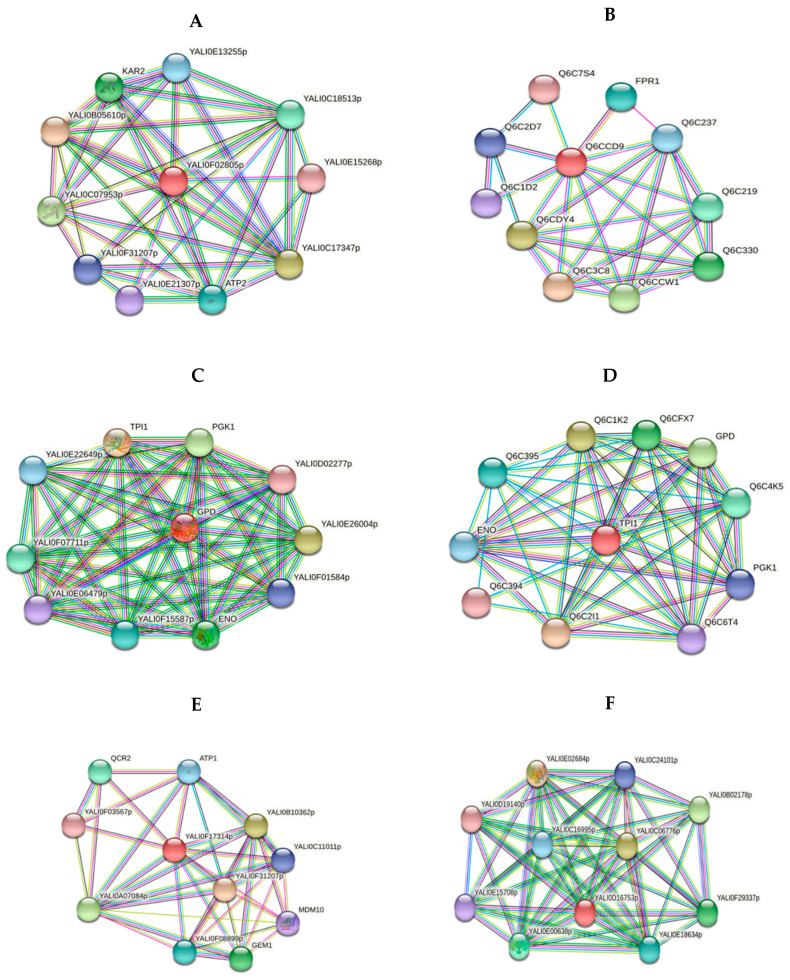
Figure 4.
Network analyses of functional associations and/or interactions of up-regulated proteins upon the adaptation to alkaline stress. Different clusters of interacting proteins were identified using the STRING software with the highest conformity score. The information on the proteins is summarized in the Supplementary Section (Table S2). (A) HSP60 heat shock protein—chaperone (YALI0F02805p) (B) Peptidyl-prolyl cis-trans isomerase (YALI0C10230p) (C) Glyceraldehyde-3-phosphate dehydrogenase (YALI0C06369p) (D) Triose phosphate isomerase (YALI0F05214p) (E) VDAC (YALI0F17314p) (F) Malate dehydrogenase (YALI0D16753p).