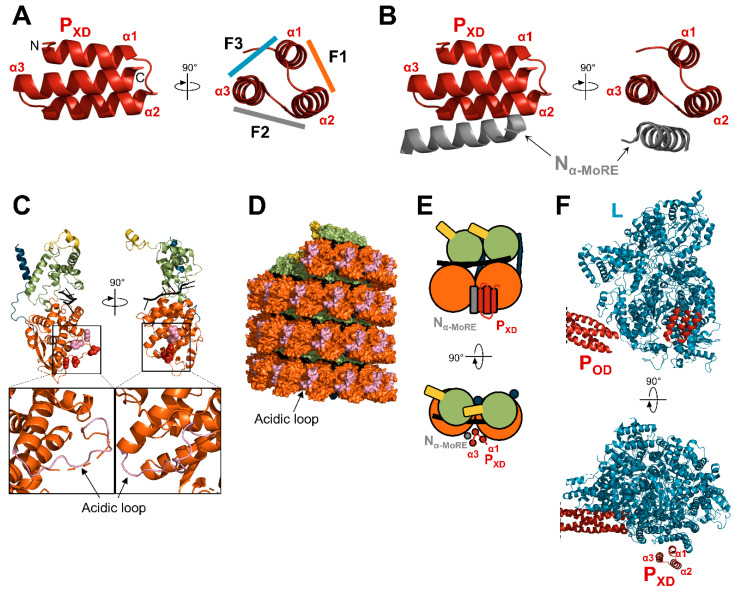
Figure 9.
Interaction network of PXD. (A) Structure of MeV PXD (PDB: 1T6O). The three faces of the “prism” are indicated (F1, F2, F3). F1, F2, and F3 are shown with a color code corresponding to the color used to draw the protein partner. (B) Structure of MeV PXD in complex with Nα-MoRE (PDB: 1T6O). (C) Structure of a protomer of MeV N, as observed in the NCLC (PDB: 6H5Q) with the RNA shown in black. Amino acids shown as red spheres correspond to CDV N residues that once mutated restore the growth of a recombinant CDV bearing deletions in the NCDR [191]. The amino acids corresponding to the residues of NiV identified as important for the interaction with PXD are shown as pink spheres [192]. Bottom: enlargements of the NNTD with the acidic loop shown in pink. (D) Surface representation of a nucleocapsid-like complex of MeV (reconstituted from PDB: 6H5Q) with the acidic loop shown in pink. (E) Schematic representation of two adjacent N protomers bound to PXD in complex with Nα-MoRE. (F) Structure of the polymerase complex of PIV5 (PDB: 6V85). For panels (C–E), N protomers are colored according to the color code in Figure 4.