Figure 1.
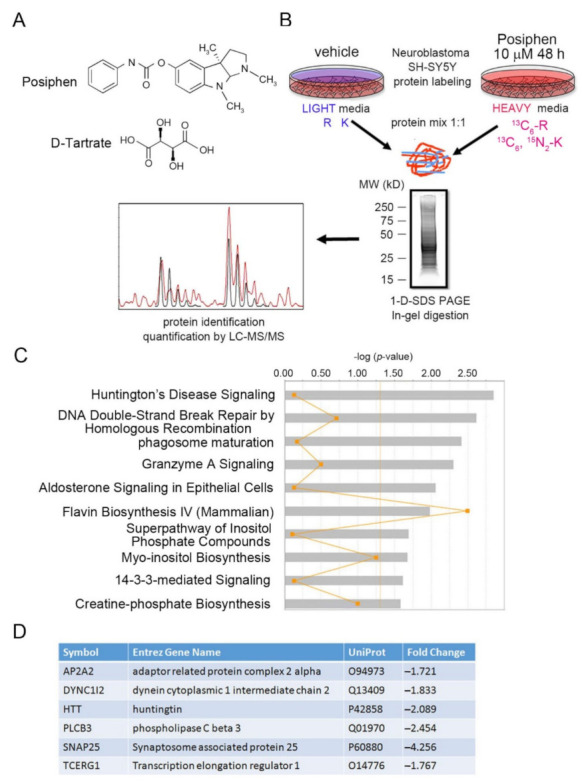
Main canonical pathways affected by Posiphen using the stable isotope labeling with amino acids in cell culture (SILAC) method for SH-SY5Y cells. (A) Chemical structure of Posiphen as the Tartrate salt. (B) Cell lysate from the two treatment conditions (with or without Posiphen 10 μM) were mixed in equal quantities and loaded on a gel. The obtained gel stained with SYPRO® is reported. Gel was cut and subjected to tryptic digestion prior to analysis by mass-spectrometry in Nano LC-MS/MS Orbitrap Q-Exactive. Raw data were subjected to bioinformatics analysis to identify and subsequently filter proteins with significant changes in expression levels. Ingenuity Pathway Analysis (IPA) identified cluster pathways affected by the treatment. MW: molecular weight; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; LC-M/MS: liquid chromatography tandem mass spectrometry. (C) Differentially expressed proteins were clustered by IPA. The 10 most statistically significantly affected (p < 0.05) canonical pathways are shown. The significance threshold value (-log [p-value]), set at 1.3, is highlighted in orange. Orange pints are the ratio of the number of genes that meet the cut-off criteria/the number of genes that make up that pathway. (D) The most downregulated proteins (≤1.7) in the Huntington’s pathway are listed.
