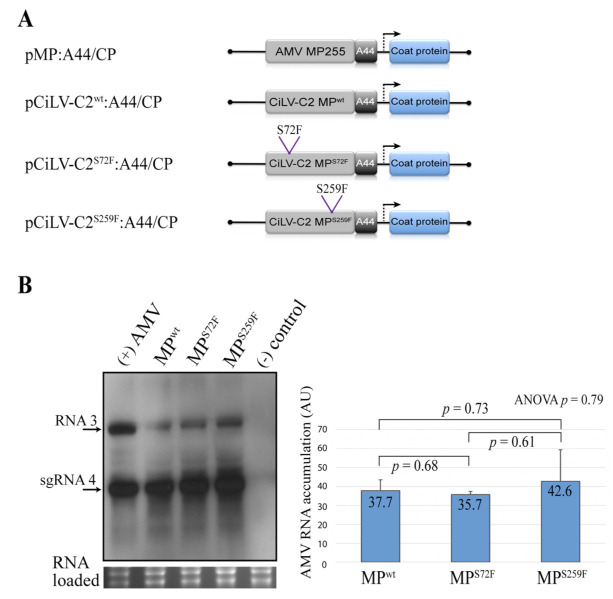
Figure 4.
Northern blot analysis of the accumulation of AMV RNA in P12 protoplasts. (A) The schematic representation shows the pAL3NcoP3 AMV RNA 3 construct [36], lacking the GFP gene and its derivatives carrying the heterologous CiLV-C2 MPs wild type (WT) and mutated versions (MPS72F and MPS259F) fused to the C-terminal 44 residues of the AMV MP (A44), used in this assay. (B) Northern blot showing accumulation of AMV RNAs in P12 protoplasts at 16 h post-inoculation, using a dig-riboprobe complementary to the 3′ UTR of the AMV RNA 3. Transcripts correspond to the AMV RNA 3 derivative carrying the CiLV-C2 MPWT, mutated (MPS72F and MPS259F) and AMV MP (positive control) proteins. Negative control (-) corresponds to the non-infected P12 protoplasts. The localization of RNA 3 and subgenomic RNAs (sgRNA 4) are indicated. The graph represents the total accumulation of AMV RNA, carrying the constructs CiLV-C2 MPWT, MPS72F, and MPS259F from the average of two independent experiments. The bands were quantified using the ImageJ v. 2.0cr software with the ISAC plugin and error bars represent standard deviation. Statistical analyses were performed using Student’s t-test and one-way ANOVA. Significant differences are interpreted for p values ≤ 0.05. p-values were obtained from pairwise comparisons between treatments or among the three groups.