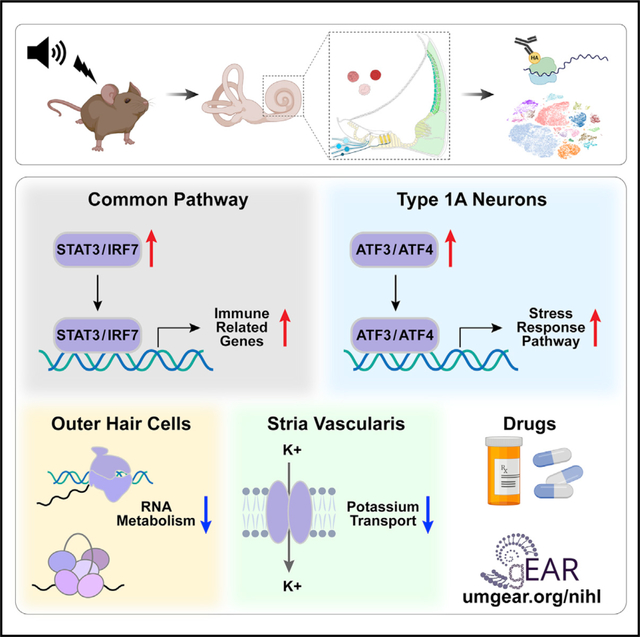
SUMMARY
Noise-induced hearing loss (NIHL) results from a complex interplay of damage to the sensory cells of the inner ear, dysfunction of its lateral wall, axonal retraction of type 1C spiral ganglion neurons, and activation of the immune response. We use RiboTag and single-cell RNA sequencing to survey the cell-type-specific molecular landscape of the mouse inner ear before and after noise trauma. We identify induction of the transcription factors STAT3 and IRF7 and immune-related genes across all cell-types. Yet, cell-type-specific transcriptomic changes dominate the response. The ATF3/ATF4 stress-response pathway is robustly induced in the type 1A noise-resilient neurons, potassium transport genes are downregulated in the lateral wall, mRNA metabolism genes are downregulated in outer hair cells, and deafness-associated genes are downregulated in most cell types. This transcriptomic resource is available via the Gene Expression Analysis Resource (gEAR; https://umgear.org/NIHL) and provides a blueprint for the rational development of drugs to prevent and treat NIHL.
In brief
Milon et al. show that cell-type-specific transcriptomic changes following noise exposure dominate the response compared to common changes. The noise-resilient type 1A neurons induce the ATF3/ATF4 stress-response pathway, and the outer hair cells and lateral wall downregulate mRNA metabolism genes and potassium transport genes, respectively.
Graphical Abstract
INTRODUCTION
Hearing loss afflicts nearly 1.57 billion people worldwide, and the incidence is expected to rise as a result of the increase in life expectancy and recreational noise exposures (Cederroth et al., 2013; GBD 2019 Hearing Loss Collaborators, 2021). The mammalian inner ear houses the cochlea, the intricate organ of hearing, which is comprised of at least four distinct functional domains (sensory epithelium, neuronal compartment, lateral wall [LW], and immune cells; Figure 1A). The sensory epithelium consists of mechanosensitive inner and outer hair cells (IHCs and OHCs, respectively), as well as a heterogeneous population of supporting cells (SCs). IHCs transmit sensory information to neurons via glutamatergic ribbon synapses, while OHCs primarily amplify and sharpen the sound stimulus (McPherson, 2018). SCs participate in ion recycling, structural support and repair, and are akin to the glial cells in the brain (Monzack and Cunningham, 2013; Wan et al., 2013). The spiral ganglion neurons (SGNs) consist of type 1 and 2 neurons, which transmit information from the IHCs and OHCs, respectively. Based on threshold and spontaneous rate, type 1 SGNs are further subclassified into three types (1A, 1B, and 1C), where type 1C are lost following noise exposure and aging and are believed to be important for hearing in the presence of competing noise (Shrestha et al., 2018; Sun et al., 2018). The LW consists of the stria vascularis (SV) and the spiral ligament. The SV contains three cell types (marginal, intermediate, and basal), which generate and maintain the endocochlear potential (EP) by secreting potassium ions into the endolymph (Wangemann, 2002). The spiral ligament consists of three cell types (perivascular endothelial cells, root cells, and fibrocytes), and together with the SV, function to maintain the EP. Finally, immune cells play a role in the inner ear response to damage (Warchol, 2019).
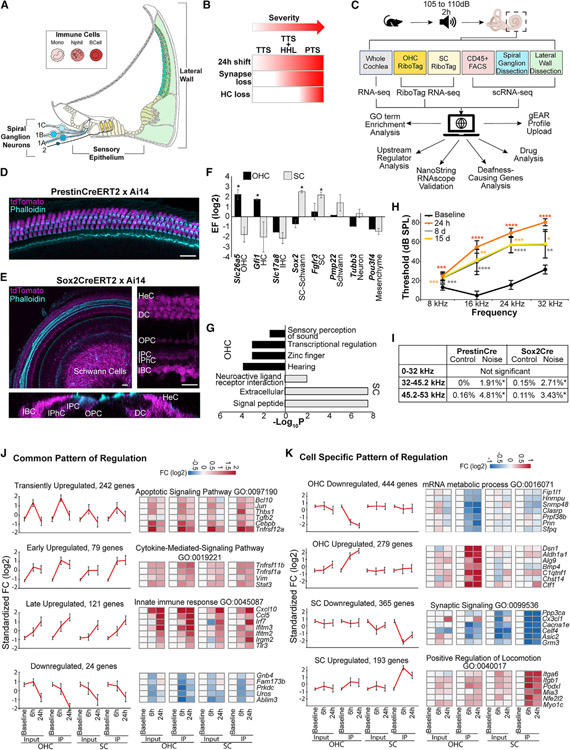
Figure 1. Experimental design and response of OHCs and SCs to noise.
(A) Schematic representing the different domains of the cochlea addressed by the experimental design.
(B) The long-term cellular and functional consequences of noise exposure present as a continuum with compounding effects.
(C) Schematic of the experimental design.
(D and E) Confocal images of whole mounted P14 mouse cochlear ducts. A cross of Prestin-CreERT2 with Ai14 (n = 5) validates the specific expression of the Cre recombinase in OHCs (D). A cross of Sox2-CreERT2 with Ai14 (n = 4) indicates recombination of the Cre in Deiters’ cells (DCs), inner pillar cells (IPCs), outer pillar cells (OPCs), inner phalangeal cells (IPhCs), inner border cells (IBCs), Hensen’s cells (HeCs), and in the glial cells. Bottom panel is an orthogonal view of a Z stack showing the absence of staining in the hair cells. Scale bar, 20 μm.
(F) Enrichment and depletion of cell-type-specific markers in the RiboTag-IP when compared to the input (n = 2 paired input-IP for each dataset). Asterisk (*) indicates genes meeting the enrichment criteria described in the methods.
(G) Overrepresented GO categories among genes with expression specifically enriched in SCs or OHCs (p values calculated using hypergeometric test).
(H) ABR thresholds from 9-to-11-week-old mice (n = 15) exposed to 105 dB SPL for 2 h before and 24 h, 7 days, and 15 days post noise exposure. Error bars: standard deviations. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Two-way ANOVA with Tukey’s post hoc test.
(I) Percentage of missing OHC along different frequency ranges. (PrestinCre: n = 3 biological replicates for each condition; Sox2Cre: n = 6 controls, n = 5 noise-exposed). *p < 0.05. Unpaired t test.
(J and K) Integrated analysis of the OHC and SC datasets delineates the transcriptional responses to noise that are common (J) and OHC- or SC-specific (K). Error bars: standard deviations. Selected genes from each cluster are shown using heatmaps.
Exposure to sustained loud noise results in an initial increase in the hearing thresholds, which may completely or partially reverse over a period of days to weeks, resulting in a temporary or permanent threshold shift (TTS or PTS, respectively) (Hertzano et al., 2020a; Ryan et al., 2016). The long-term cellular and functional consequences of noise exposure present as a continuum. This ranges from no structural damage in a TTS, to a TTS with axonal retraction of the type 1C SGNs (also known as hidden hearing loss) (Liberman and Kujawa, 2017), to permanent damage associated with dysfunction and/or loss of hair cells (primarily OHCs) in a PTS-type injury (Figure 1B). Additional changes have been reported in the SV, spiral ligament, and immune response (Herranen et al., 2018; Hirose and Liberman, 2003). The progressive loss of OHCs and SGNs also account for the pathophysiology of age-related hearing loss (ARHL) (Wu et al., 2020). Therefore, noise exposure paradigms are used to screen therapeutics for both conditions. However, to date, there are no treatments to reverse or prevent noise-induced hearing loss (NIHL) or ARHL (Schilder et al., 2019).
Targeted therapeutics require a detailed molecular understanding of the observed pathophysiology. Yet, the structural complexity of the mammalian inner ear compounds the interpretation of bulk RNA sequencing (RNA-seq) experiments designed to reveal molecular changes induced by noise exposures (Cederroth et al., 2019; Jongkamonwiwat et al., 2020; Maeda et al., 2021). Because of the cellular diversity of each of the functional subdomains of the cochlea, maximal insight is gained when utilizing a cell-type-specific approach, such as single-cell RNA-seq (scRNA-seq), to analyze each subdomain separately (Hoa et al., 2020; Kolla et al., 2020; Korrapati et al., 2019; Petitpré et al., 2018; Shrestha et al., 2018; Sun et al., 2018). Yet, OHCs and SCs of the mature inner ear are particularly difficult to dissociate due to their robust actin cytoskeleton and tight junctions (Burns et al., 2013), which hinder their examination by current scRNA-seq protocols. Thus, transcriptomic analysis of adult OHCs and SCs benefits from alternative approaches, such as cell-type-specific immunoprecipitation of ribosomes (e.g., RiboTag) (Chessum et al., 2018; Hertzano et al., 2020b; Sanz et al., 2009).
In this study, we used a combination of RiboTag and scRNA-seq to generate a comprehensive, cell-type-specific molecular blueprint of the adult mouse inner ear, both before and after PTS-inducing noise exposure (Figure 1C). Through this combinatorial approach, we identified gene expression signatures that are specific to the various cell types in each of the cochlear functional domains, as well as a common molecular response to PTS-inducing noise, which is likely driven by the transcriptional regulators STAT3 and IRF7. We further analyzed this dataset to propose signaling pathways across cell types in response to noise, identify noise-induced changes in the expression of deafness-causing genes, and discover candidate therapeutics to prevent/ameliorate NIHL or ARHL. Finally, we made this rich resource available for browsing and analysis via the Gene Expression Analysis Resource (https://umgear.org/NIHL).
RESULTS
Distinct and shared responses of OHCs and SCs to PTS-inducing noise exposure
To investigate the molecular response of OHCs and SCs to PTS-inducing noise, we used the RiboTag approach, which enriches for cell-type-specific actively translated genes without tissue dissociation (Sanz et al., 2009). By sequencing both the input (whole cochlea) and the cell-type-specific immunoprecipitated RNA (RiboTag-IP), comparative analyses can delineate cell-type-specific transcriptional programs and responses to stress (Hertzano et al., 2020b; Sadler et al., 2020). In the mature inner ear, prestin is the canonical marker of OHCs (Zheng et al., 2000), whereas Sox2 is expressed in all SCs (Walters et al., 2015). We crossed RiboTag mice with Prestin-CreERT2 (Fang et al., 2012) and Sox2-CreERT2 (Arnold et al., 2011) mouse strains to enrich for OHC and SC transcripts, respectively. To test the specificity of recombination, both Cre lines were also crossed with the ROSA26CAG-tdTomato (Ai14) mice for evaluation by immunofluorescence (Madisen et al., 2010). Following sequencing, we assessed the enrichment and depletion of known cell-type-specific transcripts. As expected, Prestin-CreERT2 induced recombination in all OHCs and resulted in the enrichment of OHC-expressed transcripts along with depletion of non-hair cell expressed genes (Figures 1D and 1F). Conversely, the Sox2-CreERT2 induced recombination in SCs (medial SCs, pillar, and Deiters’) as well as within a population of cells in the spiral ganglion, identified as Schwann cells, based on the enrichment of Pmp22 in the RNA-seq results (Figures 1E and 1F). To further establish the utility of this model system, we compared gene expression levels between the RiboTag-IP and input samples and detected 436 and 248 genes specifically enriched for OHCs and SCs, respectively (Table S1). Gene Ontology (GO)-term analysis showed that hearing-related genes were overrepresented in the set of OHC-enriched transcripts (including the deafness genes Espn, Kcnq4, Loxhd1, Slc26a5, Strc, Tmc1, Tomt, and Smpx), while the SC-enriched genes showed significant overrepresentation of genes that function in neuronal-related activities (Figure 1G).
To characterize the response to noise in OHCs and SCs, adult mice were exposed to an 8 to 16 kHz noise band at 105 dB SPL for 2 h, resulting in a PTS across all frequencies (Figures 1H and 1I; Figure S1A). We measured gene expression at 6 h and 24 h after noise exposure, as well as in unexposed controls. Differential expression (DE) analysis was used to identify genes with a significant change in expression in the RiboTag-IP samples after noise in comparison to the unexposed controls (FDR < 5% and fully separated expression levels between conditions). 1,947 and 1,796 genes were detected as differentially expressed in the OHC and SC datasets, respectively (Table S1). To define the main cell type and kinetic response patterns detected in this experiment, we next integrated these two datasets and subjected the union of the noise-responding genes to a clustering analysis. We contrasted the response measured in the OHC and SC IP samples with that measured in the entire cochlea (input samples) to delineate specific and shared responses (Table S2).
This analysis identified four major clusters that had a similar kinetic pattern across all samples (Figure 1J; Table S2), likely representing a more general core response to noise. GO-term analysis found that the cluster of transiently induced genes is enriched for regulators of apoptosis (including Jun, Bcl10, and Tgfb2), and the cluster containing genes with a stronger induction at 24 h is enriched for immune-related genes (including Ifr7, Ccl5, and Tlr3). Conversely, our analysis identified four clusters that manifested a response specific to OHC or SC RiboTag-IP samples (Figure 1K) that likely represent cell-type-specific responses. GO-term analysis showed that the cluster of genes specifically repressed in OHC RiboTag-IP samples was enriched for genes that function in mRNA metabolic processes (e.g., Sfpq, Hnrnpu, and Snrrnp48). In contrast, the cluster of genes that were specifically repressed in SCs was enriched for genes that function in transmission of nerve impulse (e.g., Grm3, Asic2, and Cacna1e), while the cluster of genes that were specifically induced in the SC RiboTag-IP samples was enriched for genes encoding structural proteins involved in cell adhesion and migration (e.g., Podxl and Itgb1) (Figure 1K; Table S2). Finally, we selected 65 genes enriched in the Ribotag-IP of either or both datasets for validation in independent replicates, including regulators of mRNA metabolism and synaptic signaling-related genes. NanoString validated the change in gene expression for 43 genes following noise exposure (Figures S1B–S1D; Table S2).
Induction of the ATF3/4 pathways and repression of genes involved in neuronal transmission in the type 1A SGNs following noise
Sustained exposure to loud noise results in glutamatergic excitotoxicity, leading to the retraction of type 1C SGN axon nerve terminals, also known as synaptopathy. To enhance our understanding of the SGNs’ common and cell-type-specific responses to noise, we compared the SGN transcriptomes of adult mice 24 h after PTS-inducing noise exposure with unexposed controls using scRNA-seq (Figure S1E). Utilizing a dissection procedure optimized to enrich for sensory neurons, we obtained tissue from four biological replicates per condition and processed for sequencing using a droplet-based microfluidics platform. We merged the control and noise-exposed datasets using anchor-based data integration (a computational method that builds on the identification of multiple pairs of cells where each pair is composed of one cell from each dataset, which are mutually most similar to each other [“anchors”]. Such anchors likely represent cells in the same biological state across the analyzed datasets and therefore can guide their merge) (Stuart et al., 2019). Based on the expression of known marker genes, distinct cell clusters were identified for types 1A (Calb2), 1B (Calb1), 1C (Lypd1), and type 2 (Prph) SGNs (Hafidi, 1998), as well as for Schwann cells (Pmp22) (Amici et al., 2006) (Figures 2A and 2B; Table S3). Collectively, these clusters, consisted of 8,916 cells out of the 25,994 cells in this dataset (Figures S2A–S2E). UMAP visualization of the integrated dataset confirmed that cell types preserved their identity after noise exposure (Figure S2F) and that cells did not group according to individual replicates (Figure S2G). Furthermore, marker genes showed similar expression patterns in cells from control and noise-exposed samples (Figures S2H–S2J; Table S3), corroborating that the analysis properly captured cell-type identities in both conditions. In agreement with the expected SGN composition, 96% and 4% of the neurons were assigned as type 1 and type 2, respectively (Perkins and Morest, 1975; Ryugo and Parks, 2003). Type 1 SGNs were further divided into 28%, 32%, and 40% 1A, 1B, and 1C subtypes, respectively. These different cell types showed similar prevalence in the control and noise-exposed samples (Figure S2K).
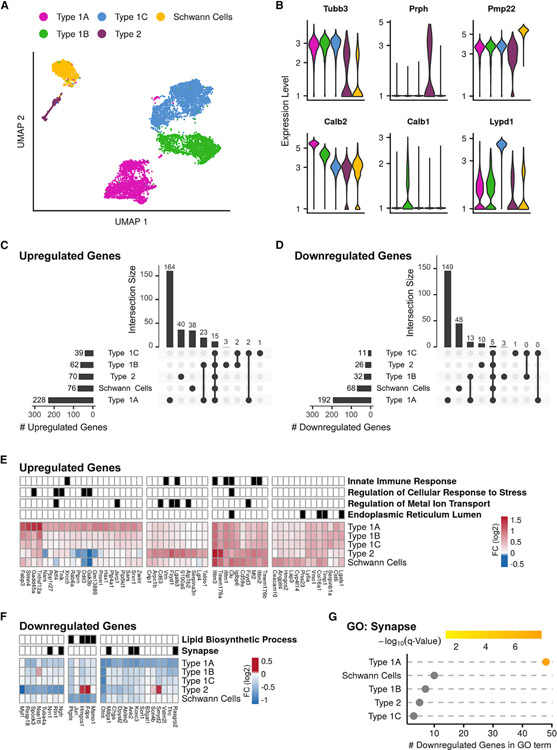
Figure 2. scRNA-seq analysis of the response of the SGNs to PTS-inducing noise exposure.
(A) UMAP of the 8,916 SGNs and Schwann cells (n = 4 biological replicates for each condition).
(B) Violin plots for the expression of known marker genes, colored according to cell type as in (A).
(C and D) Upset plots of upregulated (C) and downregulated (D) DEGs in noise-exposed versus control samples. Horizontal bars: overall number of DEGs detected in each cell type. Vertical bars: number of DEGs in selected intersections between cell types indicated below the bars.
(E) Hierarchical clustering applied to the set of 56 upregulated genes that showed, upon noise exposure, a fold-change induction greater than 1.5 and FDR q value <0.05 (MAST’s statistical test) in at least one of the cell types. The grid above the heatmap displays assignment of genes to enriched GO terms (q < 0.05, hypergeometric test) (Table S3).
(F) Same as (E) but for the 27 downregulated genes (Table S3).
(G) The GO term “synapse” is enriched in the downregulated DEGs of type 1A. (q < 0.05, hypergeometric test).
Next, for each separate cell type, we performed DE analysis comparing noise-exposed and control cells. Collectively, this analysis identified 321 and 255 differentially expressed genes (DEGs; FDR < 5%; absolute fold change > 1.2) that were upregulated and downregulated, respectively, upon noise exposure (Table S3). The majority of these DEGs met the DEG-calling criteria in only a single cell type: 76% (246) of the upregulated and 82% (211) of the downregulated genes (Figures 2C and 2D). Markedly, type 1A SGNs showed the strongest response to noise (228 and 192 up- and downregulated genes, of which 164 and 149 genes were called only in this cell type) (Figures 2C and 2D). A similar number of cells were assigned to type 1A, 1B, and 1C SGNs (Figure S2K), demonstrating that the significantly higher number of DEGs detected for type 1A is not a mere result of higher detection power for this cell type. 86 and 50 DEGs were specifically detected in Schwann cells and type 2 SGNs, respectively, while only 8 DEGs were specific to type 1B and 1C SGNs combined. Clustering the DEGs by their response pattern further delineated cell-type-specific and shared responses (Figures 2E and 2F). The sets of genes that were specifically induced in type 1A, type 2, and Schwann cells were enriched for genes that function in cellular responses to stress, metal ion transport, and the endoplasmic reticulum lumen, respectively. Genes that showed a shared induction across cell types were enriched for innate immune genes (Figure 2E; Table S3). Genes that were specifically repressed in type 1A were enriched for genes involved in synapse (e.g., Ank2, Kcnc3, and Rasgrp2; Figures 2F and 2G) as well as the GO term “glutamatergic synaptic transmission” (e.g., Gria2 and Grin1).
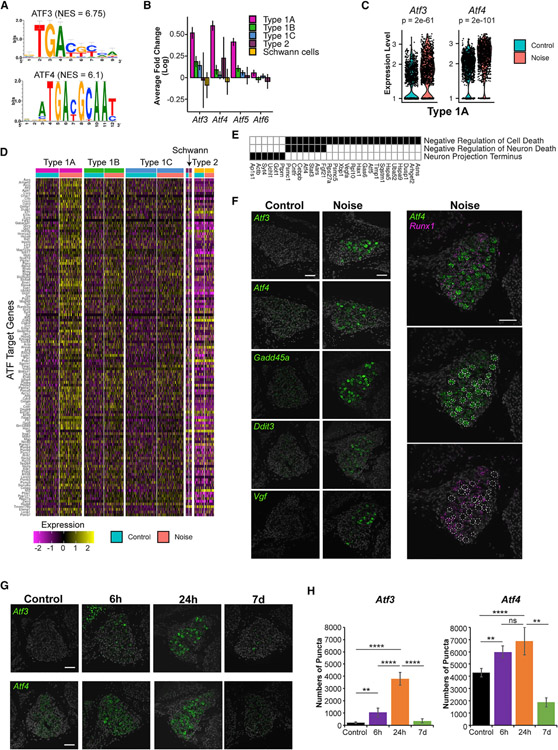
The strong noise-induced transcriptional response elicited by type 1A SGNs, in contrast to a much weaker response shown by type 1C, was unexpected, considering that type 1A SGNs are largely resilient to PTS noise while type 1C are highly susceptible (Furman et al., 2013; Shrestha et al., 2018). Seeking key regulators of the observed transcriptional response, we applied a cis-regulatory motif analysis to the promoters of the DEGs. Notably, this analysis revealed that the promoters of the genes induced in type 1A SGNs are significantly enriched for the binding motif of the ATF family of transcription factors (TFs) (Figure 3A; Table S3), pinpointing members of this family as major mediators of the type 1A SGN response to noise. Corroborating such a role, the Atf3, Atf4, and Atf5 genes themselves showed a markedly high induction in type 1A SGNs in response to noise exposure (Figures 3B and 3C). Of note, Atf3 and Atf4 are known to be induced in neurons following multiple stresses and are associated with neuronal survival and axonal regeneration (Hunt et al., 2012; Sun et al., 2013). Consistent with this finding, the 125 genes induced in type 1A and predicted as direct ATF targets by the motif analysis (Figure 3D) were enriched for the “negative regulation of cell death” GO term (including pro-survival genes like Fgf21, Cntfr, Hspa5, and Gas6; Figure 3E), suggesting that this pathway confers a protective role in type 1A cells upon noise exposure. We selected key regulators and effectors in the ATF-mediated response network (Atf3, Atf4, Gadd45a, Ddit3 and Vgf) and validated their activation in response to noise (Figure 3F). Co-staining of noise-exposed spiral ganglia with probes for Atf4 and Runx1—a marker for the type 1B and 1C SGNs (Figure S2J)—showed non-overlapping patterns of induction consistent with the induction of Atf4 in type 1A SGNs only (Figure 3F). Analysis of the ATF-induced transcriptomic response at baseline, 6 h, 24 h, and 7 days post-exposure showed a maximal response at 24 h with a near-complete resolution by 7 days (Figures 3G and 3H). Staining for Atf3 and Atf4 in the SV and the organ of Corti reveals that in these cochlear domains, in contrast to the SGNs, the peak of expression following noise is at 6 h (Figures S3A and S3B). Furthermore, while both Atf3 and Atf4 expression is highly induced in the SGNs and the basal cells of the SV (Figure S3A), only Atf4 is detected and induced in the organ of Corti in a diffuse pattern of expression (Figure S3B).
Figure 3. ATF transcription factors regulate the type 1A transcriptional response to noise.
(A) Top scoring enriched motifs on promoters of the genes induced in type 1A SGNs. NES, normalized enrichment score.
(B) Several genes from the Aft family display a greater induction in type 1A compared to other SGNs. Error bar: 95% confidence interval from MAST DE analysis.
(C) Violin plots showing Atf3 and Atf4 expression in type 1A control and noise-exposed cells. p values calculated using MAST.
(D) Heatmap of normalized and scaled expression levels of ATF-predicted target genes induced by noise in type 1A SGNs. Rows are genes and columns are cells.
(E) Selected GO terms enriched in the ATF-predicted targets from (D) (Table S3).
(F) Representative images of RNAscope labeling for selected ATF-target transcripts in the spiral ganglia of control and noise-exposed mice show an increase in gene expression in type 1A SGNs following noise (n = 3 controls, n = 3 noise-exposed). Scale bar, 50 μm.
(G) Time course of Atf3 and Atf4 induction following noise exposure. Scale bar, 50 μm.
(H) Quantitative analysis of the RNAscope labeling for Atf3 and Atf4 using QuPath (see Method details). Error bars: standard deviation (n = 3 biological replicates for each condition). **p < 0.01; ****p < 0.0001; ns not significant. One-way ANOVA with Tukey’s post hoc test.
An analysis focused on the downregulated genes detected a weaker, albeit significant, enrichment for the binding signature of the transcription factor SREBF1 in 39 of the type 1A downregulated genes (Figures S3C–S3F). These genes overrepresented GO terms related to neuronal transmission, such as synapse and neuron projection (Figure S3G; Table S3). Here, too, Srebf1 itself showed a greater suppression of expression upon noise exposure in type 1A cells compared to other SGN cell types (Figures S3D and S3E), supporting a role for its attenuated activity in mediating the suppressed transcriptional program observed in noise-exposed type 1A SGNs.
LW cells downregulate genes related to potassium transport following noise exposure
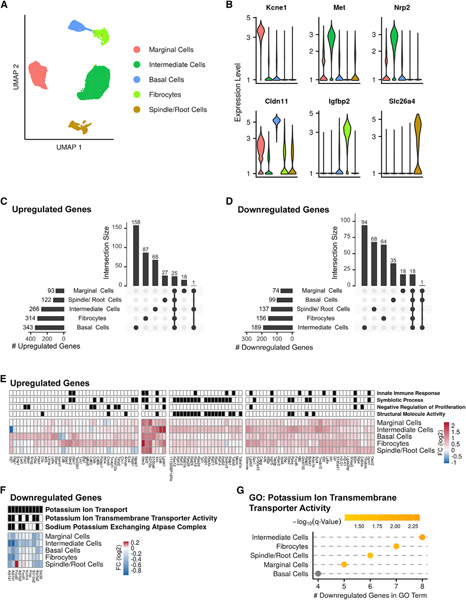
The cochlear LW consists of the SV and spiral ligament, which function to maintain the EP necessary for the mechanotransduction of sound. The SV consists of three layers of cells (from medial to lateral: marginal, intermediate, and basal) that secrete potassium ions into the endolymph of the scala media via a network of Na+/K+ transporters (Peixoto Pinheiro et al., 2021). Importantly, the marginal and intermediate cells of the SV and the fibrocytes in the spiral ligament have been shown to endure structural changes following PTS-inducing noise exposure (Hirose and Liberman, 2003). Using the same scRNA-seq strategy as before, we explored the noise-induced transcriptional responses of the LW to understand the molecular underpinnings of SV and spiral ligament pathology following PTS-inducing noise. We clustered the 34,341 cells that met quality criteria and used the expression of known marker genes to identify clusters corresponding to five of the major cell types in the LW— marginal cells, intermediate cells, basal cells, fibrocytes, and spindle/root cells—and re-clustered the 25,599 cells assigned to these five cell types (Figures S4A–S4G). As in the SGN dataset, control and noise cells were integrated using an anchor-based integration approach. The UMAP showed well-separated clusters that were markedly enriched for the marker genes of the respective cell types (Figures 4A and 4B; Figure S4I; Table S4) (Korrapati et al., 2019). The different cell types detected in the LW showed similar prevalence in the noise-exposed and control samples (Figure S4H). SV epithelial cells (marginal, intermediate, and basal) represented 87% of the cells of the LW, with intermediate cells being the most abundant cell type (~50%) in this dataset (Figure S4H).
Figure 4. scRNA-seq analysis of the response of the lateral wall to noise.
(A) UMAP of the 25,599 LW cells (n = 4 biological replicates for each condition).
(B) Violin plots for the expression levels of known marker genes, colored according to cell type as in (A).
(C and D) Upset plots of upregulated (C) and downregulated (D) DEGs in noise versus control comparisons.
(E) Heatmap showing the DEGs induced by a fold change greater than 1.5 (FDR q < 0.05; MAST’s statistical test) in at least one cell type. The grid above the heatmap shows selected GO term enriched in each of the clusters (q < 0.05, hypergeometric test).
(F and G) A program of gene repression upon noise exposure shared by most of the LW cell types is enriched for genes that function in potassium transport (hypergeometric test).
We next performed, for each separate cell type, a DE analysis comparing noise-exposed and unexposed controls. Collectively, we detected 639 upregulated and 408 downregulated DEGs, out of which 356 and 279 were called in a single cell type (Figures 4C and 4D; Table S4). Clustering the DEGs that showed the strongest induction (fold change [FC] of at least 1.5) delineated both cell-type-specific and shared responses: genes specifically responsive in intermediate cells were enriched for genes associated with disease process such as anemia, leukemia, and migraines, while the shared response was enriched for innate immune response (Figure 4E; Table S4). Notably, a downregulation response that was shared by most LW cell types was significantly enriched for potassium ion transport genes (Figures 4F and 4G), reflecting the deterioration, upon noise exposure, in the functioning of the LW in transporting potassium into the endolymph.
Monocytes are the primary immune cell to transcriptionally respond to PTS-inducing noise
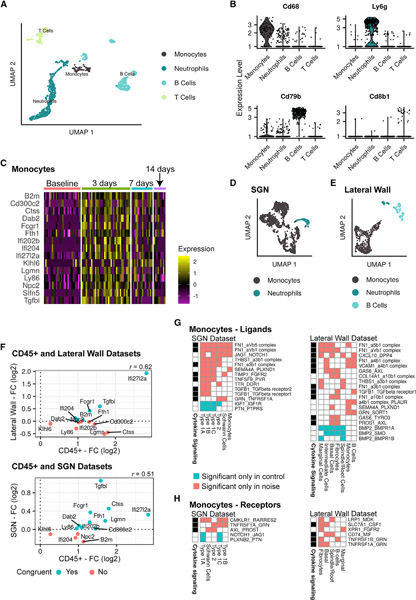
Our data show a robust induction of immune-related genes in the cochlear sensory epithelium (OHCs and SCs), LW, and SGNs in response to PTS-inducing noise. To directly measure the molecular response in the cochlear immune cells following noise exposure, cochleae were obtained at three time points after noise exposure (3, 7, and 14 days), as well as from control mice (Figure S1F). Flow cytometry was used to select CD45+ immune cells for scRNA-seq analysis (Figure S1G). UMAP visualization of the 1,123 cells, which met quality criteria from all four conditions, indicated that cells clustered according to their identity rather than experimental condition (Figures S5A–S5F). Expression of canonical marker genes for the major immune cell types defined clusters of B cells, T cells, monocytes/macrophages, and neutrophils (Figures 5A and 5B; Figures S5G and S5H; Table S5). In this dataset, 47% of the cells were identified as neutrophils, while 30%, 15%, and 8% were identified as B cells, T cells, and monocytes/macrophages, respectively (Figure S5F). In line with recent reports, we observed an increase in neutrophils 3 days after noise exposure (Bae et al., 2021; Rai et al., 2020).
Figure 5. scRNA-seq analysis of the response of inner ear CD45+ immune cells to PTS noise exposure.
(A) UMAP of 1,123 cochlear immune cells taken from control mice and noise-exposed mice at 3, 7, and 14 days after exposure (n = 1 biological replicate consisting of pooled tissue from 6 mice at each time point).
(B) Violin plots for the expression of known marker genes for B cells, T cells, monocytes, and neutrophils.
(C) Heatmap showing the expression levels of the 15 DEGs detected in monocytes. Columns represent cells, and color indicates scaled normalized expression levels.
(D) UMAP of 1,499 immune cells taken from control and 24 h post noise exposure in the SGN dataset.
(E) UMAP of 655 immune cells taken from control and 24 h post noise exposure in the LW dataset.
(F) Correlation between the response to noise of the DEGs detected in monocytes of the CD45+ sorted dataset and the response in monocytes from the LW and SGNs datasets. Shown are the 13 DEGs (out of 15 DEGs) whose expression was detected also in monocytes of the LW/SGNs dataset. Note: time point posttreatment differs between the two datasets: 24 h in the LW and SGN compared to 3 days in the CD45+ dataset. r, Pearson correlation coefficient.
(G) Ligand-receptor interactions detected by CellPhoneDB (FDR < 5%). Shown are interactions called either only in the noise-exposed or the control cells (and that the mean expression of the pair of ligand receptors was higher under that condition) and are mediated by a ligand secreted by monocytes. Each interaction is labeled by the pair of the ligand and receptor symbols with the ligand indicated first. The genes involved in these interactions were enriched for cytokine signaling (q = 0.003 and 0.0007 for SGNs and LW, respectively; hypergeometric test).
(H) Similar analysis to (G), focusing on receptors in the immune cells and ligands in the target cells.
Next, for each of these separate cell types, we carried out DE analysis, comparing cells from each of the three post-exposure time points to the unexposed control cells. Notably, all but two DEGs (detected for 3-day neutrophils and 7-day T cells) were exclusively detected for monocytes and only for the 3-day time point, even though, in terms of statistical power, monocytes were less abundant than the other immune cells (Figure S5F). Overall, this analysis detected 15 DEGs, all upregulated (Figure 5C) and enriched for the GO term “adaptive immune response” (Table S5). Some of these genes (e.g., KlhI6 and B2m) also showed an increased expression 7 days post noise exposure, though they did not pass the statistical significance level for being called DEGs. Most of these genes returned to baseline expression levels at 14 days post treatment (Figure 5C; Figure S5I; Table S5).
The observation that, out of the four immune cell types analyzed, only monocytes elicited a significant transcriptional response to noise exposure was unexpected. We therefore revisited the SGN and LW scRNA-seq datasets described above and examined the immune cells detected in them. In the SGNs dataset, we identified clusters representing monocytes and neutrophils (Figure 5D; Figures S5J–S5M; Table S5). DE analysis detected 243 upregulated and 125 downregulated genes in monocytes and only 3 upregulated and 3 downregulated genes in neutrophils (Table S5). In the LW dataset, clusters representing monocytes, neutrophils, and B cells were identified (Figure 5E; Figures S5N–S5R; Table S5), and here, too, monocytes appeared as the cell type that showed the most vigorous transcriptional response to noise with 306 and 257 upregulated and downregulated DEGs, respectively. Only 4 DEGs were identified in B cells and neutrophils (all downregulated in B cells) (Table S5). Notably, the upregulated DEGs detected in monocytes from the CD45+ immune cells dataset showed a correlated response in monocytes of both the LW and SGN datasets (Figure 5F). Overall, in these three scRNA-seq datasets, monocytes accounted for the vast majority of DEGs detected in immune cells.
To explore the potential impact of monocytes on other cell types of the inner ear mediated by cell-cell communication, we used the CellPhoneDB tool (Efremova et al., 2020). We detected an overall increase in communication between monocytes and SGN and LW cells following noise exposure. Numerous interactions involved in cell adhesion and secretion of chemoattractant molecules were detected (Figures 5G and 5H).
A common response of the cochlea to PTS-inducing noise is probably regulated by IRF7 and STAT3
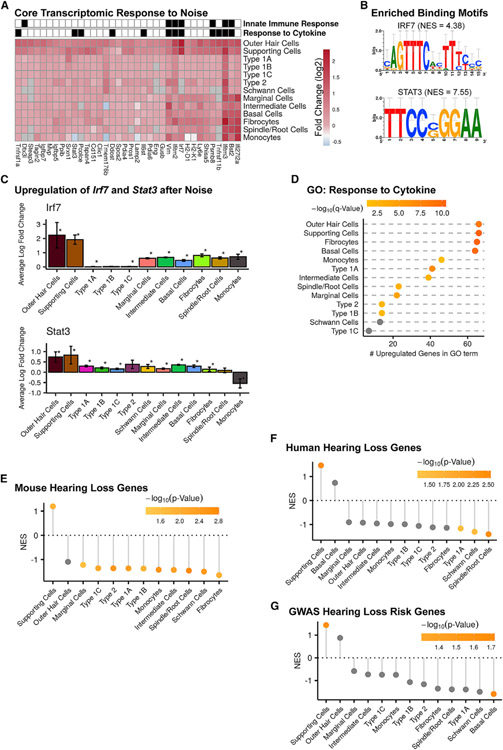
The analyses shown to this point indicate that the cochlear response to PTS-inducing noise is primarily cell-type-specific within each of the various cochlear domains, where the largest number of DEGs is specific to individual cell types rather than the domain. We next sought to identify whether the cochlea also mounts a molecular response to noise that is shared across cell types. We identified a set of 36 genes that, following noise, was upregulated in each of the three major datasets analyzed (SGN, LW, and RiboTag) in more than one cell type (Figure 6A). This set of genes represents a core transcriptional response to noise that is shared by most cells of the inner ear. Seeking key regulators of this common response, we found that the promoters of these 36 genes were significantly enriched for the binding motifs of the IRF7 and STAT3 TFs (Figure 6B). Importantly, Stat3 and Irf7 themselves were included in this set of common noise-induced genes (Figures 6A and 6C), corroborating their role as pivotal inducers of this response. From a functional standpoint, this common transcriptomic response is enriched for innate immune genes (7 genes) and cytokine pathways (12 genes) (Figure 6A). Moreover, for 11 out of 13 cell types, we found that the set of upregulated DEGs was enriched for the GO term “response to cytokine” (Figure 6D).
Figure 6. A common response to PTS-noise exposure in inner ear cells.
(A) Heatmap of the set of common upregulated genes. Gray indicates that the gene was not detected in the corresponding cell type. Shown are genes that were detected as upregulated (q < 0.05; fold change > 0; MAST’s or DESeq2 statistical test for scRNA-seq or RiboTag datasets, respectively) in at least two cell types in each of the two single-cell datasets (SGN and LW) and were members of one of the common upregulated gene clusters in the RiboTag dataset (Figure 1J). These genes were enriched for the GO terms “response to cytokine” (q = 0.0008) and “innate immune response” (q = 0.04). “Monocytes” refer to LW monocytes.
(B) The promoters of the common upregulated genes from (A) were enriched (RcisTarget analysis) for the binding motifs of IRF7 and STAT3 TFs (NES, normalized enrichment score).
(C) Upregulation of Irf7 and Stat3 transcripts was observed in most cell types in the inner ear. *q < 0.05 (MAST’s or DESeq2 statistical test for scRNA-seq or RiboTag datasets, respectively). Error bar: 95% confidence.
(D) For 11 out of 13 cell types, the set of upregulated DEGs was enriched for the GO term “response to cytokine” (q < 0.05, hypergeometric test).
(E–G) Plots showing the NES obtained from GSEA for the mouse hearing-loss-causing genes (E), for the human hearing-loss-causing genes (F), and for GWAS hearing-loss-risk genes (G) in each cell type following noise exposure. All cells, except for SCs, had a negative NES reflecting downregulation of hearing-loss genes following noise exposure.
Genes downregulated by PTS-inducing noise are enriched for deafness-causing genes
We next explored potential roles of deafness-causing genes, as well as candidate genes for ARHL from genome-wide association studies (GWAS), in NIHL. We applied a gene set enrichment analysis (GSEA) (Subramanian et al., 2005) to the main cell type in each of the four cochlear functional domains profiled in our transcriptomic assays. We tested if the genes related to hearing loss phenotypes in human or mice, when considered together as a set, show significant up- or downregulation following noise exposure. Significant modulation of expression of hearing loss genes was detected in most cell types for the mouse genes (Figure 6E; Figure S6A), in type 1A SGNs, Schwann cells, and spindle/root cells for the human genes (Figure 6F; Figure S6B), and in basal cells for GWAS risk genes (Figure 6G; Figure S6C; Table S6). Notably, in the majority of cell types, hearing loss genes showed attenuated expression following noise exposure, reflected by negative normalized enrichment scores (NESs) (Figures 6E–6G), which is consistent with most deafness-causing mutations resulting in a loss of function. SCs were the only cell type associated with a significant positive NES. While the list of mouse and human hearing loss genes is validated, the list of GWAS risk genes consists of candidate genes for ARHL, and genes identified as responsive to PTS-noise may therefore serve as candidates for validation. Given the shared pathophysiology of NIHL and ARHL, the 20 genes identified in the GWAS analysis present interesting candidates for ARHL as well.
Identification of candidate therapeutics to prevent NIHL
Critically important is the identification of new drugs to prevent and treat NIHL and ARHL. Ideal candidates would be low-cost, well-tolerated, US Food and Drug Administration (FDA)-approved, orally administered drugs. We intersected the drug-target interaction data from DrugCentral (Ursu et al., 2017) with gene expression changes identified in our combined datasets. We searched for drugs that had an opposing effect on the gene expression change induced by noise exposure. The top-ranking candidate drug to reverse molecular changes induced by noise, and therefore possibly prevent NIHL, was the anti-diabetic drug metformin (Table S7).
A cloud-based resource for visualization and analysis of the data
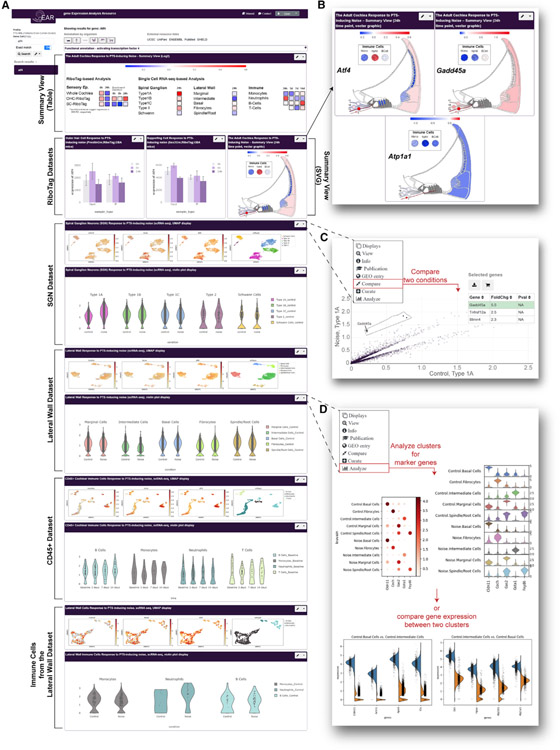
While, in accordance with the FAIR data sharing principles (Wilkinson et al., 2016), access to all data generated in this publication is provided via the Gene Expression Omnibus (Clough and Barrett, 2016), their download and interrogation require significant expertise and are time consuming, even for informatically trained individuals. To allow users, and in particular biologists, maximal and seamless usability of this resource, an innovative profile containing the data presented in this publication has been constructed in the Gene Expression Analysis Resource (https://umgear.org/NIHL; Figure 7) (Orvis et al., 2021).
Figure 7. Data sharing, visualization, and analysis via the gEAR (umgear.org).
A custom profile was generated in the gEAR to support sharing, visualization, and analysis of the processed transcriptomic data presented as part of this manuscript (https://umgear.org/NIHL).
(A) Overview of the manuscript profile, which contains two summary views (tabular and graphic, depicting gene expression as log2 fold change), the OHC and SC RiboTag datasets (bar graphs), and the SGN, LW, and immune cells datasets (UMAPs and violin plots).
(B) Examples of the graphic summary view representation for Atf4, Gadd45a, and Atp1a1 showing the (log2) fold change in gene expression following noise exposure mapped onto anatomical sites for intuitive interpretation of the data.
(C and D) The gEAR portal contains several analysis tools allowing users to further explore the data in the cloud.
(C) Example of the “compare tool,” which enables users to compare expression across any two conditions within a single dataset, here showing the DEGs in type 1A SGNs between control and noise samples.
(D) The single-cell workbench allows users to perform “de novo” analysis of the data or use a “stored analysis” to explore marker genes and compare across clusters. Here are shown marker genes for the 5 clusters (before and after noise) of the LW dataset (top) and the top 4 DEGs between intermediate and basal cells (bottom).
DISCUSSION
Understanding the molecular response of the inner ear to noise is a prerequisite for the rational development of targeted therapeutics to counter NIHL and ARHL. Here, we present a comprehensive cell-type-specific analysis of the transcriptomes of the adult mouse cochlea before and after PTS-inducing noise exposure. A major finding of our study is that within each cochlear domain analyzed, the cell-type-specific response to noise is greater than the domain-specific response. That is, within each tissue that was analyzed at the single cell level (SV, LW, and immune system), only a small number of DEGs were shared across cell types. In the LW, the shared response consisted both of the “core immune response” to noise and downregulation of a small, but likely functionally significant, cohort of potassium transport genes. Conversely, within the spiral ganglia, the shared molecular response was limited to the “core immune response” with only very few DEGs specifically detected in cells other than the type 1A SGNs.
An early adverse outcome of noise exposure is the retraction of the type 1C SGN neuronal projections from the IHCs (Coate et al., 2019; Fernandez et al., 2015), which is thought to result from glutamatergic excitotoxicity. However, rather than identifying a type-1C-specific adverse molecular response, we detected a robust molecular response of the type 1A SGNs to PTS-inducing noise exposure. This consisted of activation of the ATF3/4 pathway, which is known as the integrated stress response pathway, and depending on the severity of the trauma may result in either protective, reparative, or destructive outcomes (Pakos-Zebrucka et al., 2016). In parallel to the upregulation of the ATF pathway, type 1A SGNs show an integrated reduction of synaptic gene expression, which likely protects the neurons from overexposure to glutamate in the synapse. Indeed, several studies have shown that selectively inhibiting the glutamatergic neurotransmission by using antagonists (Duan et al., 2000; Hu et al., 2020; Kim et al., 2019; Kutzing et al., 2012; Ruel et al., 2005) or preventing the surface expression of glutamate receptors (Chen et al., 2009) prevents damage due to excitotoxicity. Similarly, downregulation of Srebp1 (activated by NMDA-receptors) has been documented as part of a protective mechanism following strokes (Taghibiglou et al., 2009). Finally, the molecular changes recorded in type 1A SGNs in this study may underlie long documented changes in neuronal physiology following acoustic trauma (Liberman and Kiang, 1978).
From a functional perspective, synaptic rearrangement (Bullen et al., 2019; Liberman et al., 2015), loss and recovery of CtBP2-labeled ribbons with juxtaposed AMPA receptors (Fernandez et al., 2015; Liberman et al., 2015), and full regeneration of lost synapses (Hickman et al., 2020; Puel et al., 1997, 1998; Ruel et al., 2007; Shi et al., 2013, 2015) have all been reported in different rodent species, suggesting that dynamic repair processes are likely an important factor in ensuring that connected neurons function properly after noise exposure. Conversely, long-term dysfunction of the synapses that remain connected after noise exposure has also been reported (Bullen et al., 2019; Shi et al., 2016), which raises the possibility that a portion of the large transcriptional responses observed in type 1A SGNs reflects an irreversible damage response. However, these findings are inconsistent with direct recordings from single SGN fibers demonstrating near-normal firing properties (Furman et al., 2013). Future experiments combining electrophysiology and molecular profiling of single neurons (e.g., using Patch-seq [Cadwell et al., 2016]) might help to disentangle this complex pathophysiology. Of note, while type 1B SGNs are also relatively protected from noise, our data suggest that this likely occurs via a mechanism that differs from the transcriptionally induced changes observed in the type 1A SGNs.
Because of the type 1C response, reversal of cochlear synaptopathy following noise has been a topic of intense research (Fernandez et al., 2021a; Hashimoto et al., 2019; Seist et al., 2020). A recently published study showed that treatment of mice with zoledronic acid after TTS-inducing noise can reverse synaptopathy, likely via the inhibition of the Farnesyl pyrophosphate synthase (FDPS) enzyme (Seist et al., 2020). Our study shows that expression of Fdps in the spiral ganglion is primarily limited to Schwann cells and type 2 SGNs, with a significant decrease in expression in Schwann cells following noise. Conversely, we detect an acute increase in the expression of Fdps in the SCs, alluding to an alternative mechanism of action or varying roles for Fdps after TTS- and PTS-inducing noise exposures. The differing expression pattern of Fdps across cochlear cell types is an example of a use case for our resource, which allows users to perform post hoc analyses of published data in addition to serving as a hypothesis generating tool.
Our data also identify a dominant molecular signature, shared by many cell types in the cochlea, of an innate immune gene-related response to noise. Our cell-type-specific approach, however, allowed for the separation of the molecular changes of immune cells from the damage-associated molecular pattern (DAMP)-related genes that are up and downregulated in non-immune cell types (Frye et al., 2019). The significance of the immune response in NIHL is still a topic of investigation. In this study, we show that the immune-related response to noise is likely driven by the transcriptional regulators STAT3 and IRF7. Indeed, inhibition of STAT3 has been shown to provide protection against NIHL through the reduction of reactive oxygen species production (Wilson et al., 2014). IRF7, part of the toll-like receptor pathway, has been shown to increase rapidly after NIHL and is thought to be part of the innate immune response to stress (Cai et al., 2014). Additionally, the acute transcriptional changes noted in monocytes/macrophages after noise trauma indicate that specific inflammatory mechanisms are involved in post-noise processes, which may be useful in developing new therapeutics.
Finally, the importance of identifying candidate therapeutics to prevent or treat NIHL cannot be overestimated. To date, there are no FDA-approved drugs to prevent or treat hearing loss. By intersecting the noise-induced gene expression changes in our combined datasets with DrugCentral (Ursu et al., 2017), we identified metformin as the top-ranking candidate therapeutic (Table S7). Indeed, metformin has been previously shown to reduce the severity of NIHL in a rat model (Kesici et al., 2018), has shown promise in prevention of ototoxicity in vitro (Oishi et al., 2014), and was shown in diabetic patients to be associated with decreased risk for sudden sensorineural hearing loss (Chen et al., 2019). Interestingly, the cholesterol lowering drugs, statins, which target HMG-CoA reductase (an indirect target of metformin), were also identified in our data as a candidate therapeutic for NIHL. Statins, similar to metformin, have shown promise in preventing NIHL (Park et al., 2012) and ototoxicity (Fernandez et al., 2021b). Finally, another high-ranking group of otoprotective candidates was halogenated compounds used as general inhalation anesthetics. Concordantly, several studies in mice have shown that treatment with isoflurane can significantly decrease threshold shifts after PTS-inducing noise (Chung et al., 2007; Kim et al., 2005). From a translational standpoint, protection by halogenated compounds may be functionally significant in otologic surgery, where the tympanic membrane and ossicular chain are manipulated in a supra-physiologic range. Taken together, these findings suggest that transcriptomic data could be used for seeking new drugs for NIHL.
LIMITATIONS AND FUTURE DIRECTIONS
Although this project represents the most comprehensive cell-type-specific analysis of the cochlear response to PTS-inducing noise, consisting of over 100,000 cells profiled and using a combination of approaches, it suffers from several technical limitations. First, the RiboTag approach, which avoids tissue dissociation that is detrimental for the robust analysis of mature hair cells and SCs, is only a method of enrichment, requiring inference of gene expression based on enrichment or depletion of the transcript compared with the input (Hertzano et al., 2020b). Furthermore, transgenic mouse models are used to drive Cre-recombination, which may also alter gene expression. Here, we used the Sox2-CreERT2 mouse to drive recombination in SCs, which also results in recombination in glial cells. However, our dissection enriched for the organ of Corti and avoided most of the glial cells, and as shown in Figure S7, the DEGs in Schwann cells and SCs are significantly divergent. Additionally, as the SCs are a heterogeneous population, our approach results in an averaged expression from the various support cell types. Finally, due to the lack of appropriate Cre-drivers, this analysis is missing IHCs, an important sensory cell that directly interfaces with the SGNs. Conversely, the single-cell analysis suffers from the disadvantage of dissociation, which may alter gene expression. It is possible that the differential expression of some acute response genes is masked by the use of a dissociation-based technique (van den Brink et al., 2017). A minor limitation is that cell-type identity is based on inference from gene expression. This mainly limits the ability to look at differences in gene expression along the tonotopic axis of the cochlea. And lastly, this dataset did not look at sex differences in the response to noise (Milon et al., 2018) and did not assess changes in protein levels or post-translational protein modification, which are not always in direct correlation to gene expression (Jongkamonwiwat et al., 2020).
Acknowledging these limitations illuminates the path for future experiments. Single-nucleus RNA-seq has evolved as a method that can efficiently replace scRNA-seq (Bakken et al., 2018; Korrapati et al., 2019). Spatial transcriptomics can be integrated with single-cell transcriptomic methods to confidently, spatially link gene expression to anatomical location (Kleshchevnikov et al., 2020). Our data indicate that, despite these advantages, multiple assays will still be ideally performed, profiling each cochlear domain separately to assess rare cell populations (e.g., type 2 SGNs, spindle cells, IHCs). Hopefully, spatial transcriptomics will reach a depth and resolution similar to standard scRNA-seq approaches and replace them as the gold standard, an advancement that will significantly reduce the cost of experiments and usage of animals. In summary, this rich resource provides users with unprecedented access to the transcriptomic cell-type-specific response to PTS-inducing noise exposure and could be used as a benchmark for molecular testing of the response of various cell types in the cochlea to new therapeutics to treat NIHL.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ronna Hertzano (rhertzano@som.umaryland.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The raw data generated during this study have been deposited in the Gene Expression Omnibus and are available under accession numbers GEO: GSE168041 (SGN and lateral wall scRNA-seq), GEO: GSE167078 (CD45+ scRNA-seq) and GEO: GSE168973 (Prestin-CreERT2;RiboTag and Sox2-CreERT2;RiboTag). All of the analyzed data are also available for browsing and analysis via the gene Expression Analysis Resource (umgear.org; https://umgear.org/NIHL).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse models
All procedures involving animals were carried out in accordance with the National institutes of Health Guide for the Care and Use of Laboratory Animals and have been approved by the Institutional Animal Care and Use Committee at the University of Maryland, Baltimore (protocol numbers 0915006, 1015003, 0918005 and 0818004), the US Army Medical Research and Materiel Command Animal Care and Use Review Office (protocol number MR130240.02), the Decibel Therapeutics Institutional Animal Care and Use Committee and the guidelines and regulations set forth by Karolinska Institutet and “Stockholm’s Norra Djurförsöksetiska Nämnd” N46/17. All mice were housed in temperature and humidity-controlled facilities with a 12-hour-light/dark cycle and with ad libitum access to food and water.
Male Ai14 mice were purchased from the Jackson laboratory (Stock No: 007914). We generated our experimental colonies by crossing and backcrossing several strains of mice that are already available. RiboTag mice (here referred as Rpl22-HA) generated by Dr. Paul S. Amieux (University of Washington) (Sanz et al., 2009) were kindly provided by Dr. Mary-Kay Lobo of the Department of Neurobiology and Anatomy at University of Maryland Baltimore. Prestin-CreERT2 knockin mice were created and kindly provided by Dr Jian Zuo of Department of Developmental Neurobiology at St. Jude Children’s Research Hospital (Fang et al., 2012). Sox2-CreERT2 mice were purchased from the Jackson Laboratory (stock No: 017593) (Arnold et al., 2011). Mice homozygous for the Sox2-CreERT2 allele are not viable and are therefore maintained as heterozygotes. CBA/CaJ mice were purchased from the Jackson Laboratory (Stock No: 000654). B6CBAF1/J mice were purchased from The Jackson Laboratory (Stock No: 100011). CBA/Ca/Sca mice were obtained from Scanbur and cryopreserved in 2011 at the Karolinska Institute Huddinge, and rederived in 2018 for this study. Crosses involving the Prestin-CreERT2 mouse model were performed as previously described (Chessum et al., 2018). Sox2CreERT2/+ were crossed to Rpl22HA/HA to generate Sox2CreERT2/+;Rpl22HA/+. This new strain was backcrossed until obtaining Sox2CreERT2/+;Rpl22HA/HA and the experimental animals were the progeny of a cross between Sox2CreERT2/+;Rpl22HA/HA with CBA/CaJ mice.
Male and female mice were used for the RiboTag experiments and male mice were used for the scRNA-seq experiments. Prestin-CreERT2;RiboTag;CBA/CaJ and Sox2-CreERT2;RiboTag;CBA/CaJ mice were 9 to 11 weeks at the time of the experiments for the RiboTag datasets. CBA/CaJ mice were 9 weeks and 7 to 8 weeks at the time of the experiments for the spiral ganglion dataset and lateral wall dataset respectively. CBA/Ca/Sca mice were 12 weeks at the time of the experiment for the CD45(+) dataset. B6CBAF1/J were 10 weeks at the time of the experiment for RNAscope.
METHOD DETAILS
Genotyping
Genotyping of Prestin-CreERT2 alleles and Sox2-CreERT2 alleles was performed by PCR with the following primers: Prestin-Fw 5′-CACAAGTTGTGAATGACCTC-3′; Prestin-Rv1 5′-TAACTGCTAGCATTTCCCTT-3′; Prestin-Rv2 5′-GTTAAAGAGCGTAATCTGGAACA-3′; Sox2-Fw 5′-TAAAGATATCTCACGTACTG-3′; Sox2-Rv 5′-TCTCTGACCAGAGTCATCCT-3′; Rpl22-Fw 5′-GGGAGGCTTGCTGGATATG-3′; Rpl22-Rv 5′-TTTCCAGACACAGGCTAAGTACAC-3′. Amplification of the prestin alleles results in a wild-type band of 300 bp and/or a Prestin-Cre band of 230 bp. The primers for the genotyping of Sox2-CreERT2 are located within the Cre recombinase gene, resulting in the amplification of 300 bp product in the presence of Cre and no amplification in the wild-type animals. Genotyping of the RiboTag alleles was performed as described previously (Sanz et al., 2009).
Induction of Cre expression
For the experiments with the Ai14 reporter mice, intraperitoneal tamoxifen injections were performed at post-natal day 6 (P6) and P7 at a dose of (3mg tamoxifen / 40 g weight). For noise exposure experiments, expression of Cre recombinase was induced by tamoxifen injections (3mg tamoxifen / 40 g weight) for two consecutive days between P10 and P15.
Noise exposure
Mice exposed to noise were between 2 to 4 months old. Mice were placed awake and unrestrained in a compartmentalized cage made of perforated aluminum sheet placed in a sound-proof chamber. Noise trauma was induced with an octave band of noise centered at 11.3kHz (8–16kHz) at 105 dB sound pressure level (SPL) for 2 hours. Sound level was measured to be within 0.5 dB of the target level throughout the holding cells, with the speaker above the mice. All mice were exposed to noise at the same time of the day (8am) to avoid introducing changes in gene expression due to circadian rhythm (Cederroth et al., 2019). For the experiments involving CD45+ immune cells, the following modifications were applied: mice were put on a rotating platform and noise trauma was induced with a broadband noise at 6–12 kHz at intensity of 110 dB SPL for 2h starting at 8 pm.
Auditory physiology
DPOAE and ABR were performed using the BioSigRP software connected to a Tucker-Davis Technologies System hardware. Mice were anesthetized using an intraperitoneal injection of a Ketamine (100mg/kg) (KetaVed Injection, Covetrus; or Ketaminol, Intervet) and Xylazine (10–20mg/kg) (AnaSed Injection, Covetrus; or Rompun, Bayer) drug cocktail. When both DPOAE and ABR were performed, DPOAE were measured first, followed by ABR. Mice were placed on a heating pad to maintain body temperature in a sound-proof chamber. Output stimuli were calibrated at the beginning of each experiment with a one-quarter inch microphone (PCB Piezotronics model PCB-378C01; or Brüel and Kjær, 4939 A 011 and 2690 A 0S1) positioned at the location where an experimental animal’s ear would be. For DPOAE, the acoustic coupler was inserted into the ear canal. A microphone (Knowles, EK 23103) inserted in the acoustic coupler with a pre-amplifier (Etymotic Research, ER-10B+) and connected to a processor (200 kHz sample rate) measured sound intensity in the ear-canal. Each speaker played one of two primary tones (f1 and f2) and declined stepwise in 5 dB from 80–10 dB SPL (for f2). To avoid distortion of no physiological origin, stimulus levels were kept ≤ 80 dB SPL. The 2f1 − f2 distortion product was measured with f2 = 8, 12, 16, 24 kHz, f2/f1 = 1.25, and stimulus levels L1 = L2 + 10 dB SPL. DPOAE thresholds were defined as the lowest level of f1 required to produce a DPOAE ≥ −5 dB SPL. Recording electrodes were inserted under the skin at the inferior postauricular area of the left and right ears, a reference was placed at the base of the skull and a ground electrode was inserted near the base of the tail. ABR were evoked by frequency-specific tone bursts 2.5 ms long (0.5 ms sinusoidal on and off ramp) at 8, 16, 24, and 32 kHz and response to each tone stimulus were recorded for 10 ms. A total of 512 sweeps were presented at the rate of 21 sweeps/s, and responses were averaged at each level and frequency tested. For each frequency, sound intensity was decreased stepwise from 90 dB SPL in 5 dB steps, until threshold was reached. Threshold was defined as the lowest sound intensity at which the reproducible waves were visually identifiable. Wave 1 amplitude was determined as the difference between the first wave peak and its subsequent trough. Baseline ABR thresholds were determined 1 week prior to noise exposure (baseline) and after the noise exposure at 24 h, 8 days, and 15 days. For the experiments involving CD45+ immune cells, the following modifications were applied: recording electrodes were placed at the head vertex (positive), under the right ear pinna (negative) and above the right leg (ground). ABRs were evoked by tone bursts 5ms long (0.5 ms rise/fall time) of 8, 12, 16 and 24 kHz, presented 33.3 times per second. Signals were collected via a low-impedance head stage (RA4LI) connected to a pre-amplifier (RA4PA) and digitally sampled with a processor (200 kHz sample rate). To determine the threshold, responses to 1000 bursts were bandpass filtered at 0.3–3 kHz using BioSigRP software and averaged. ABR and DPOAE measurements were performed at 5–7 days prior to noise trauma or sham treatment (baseline) and subsequently at sham treatment, 3 days (d), 7d and 14d post noise trauma.
Ribosome immunoprecipitation and RNA extraction
Six hours or 24 hours post-noise exposure, PrestinCreERT2/+;Rpl22HA/+;CBA and Sox2CreERT2/+;Rpl22HA/+;CBA mice (9 to 11 week-old) were euthanized by CO2 asphyxiation followed by cervical dislocation. Temporal bones were harvested, and cochlear ducts removed and immediately frozen on dry ice. Of note, while recombination occurs in supporting cells and glial cells with the Sox2-CreERT2 model, our dissection approach excludes the modiolus and thereby enriches for epithelial cells compared with neuronal tissue (as shown by the divergence in DEGs in Figure S7). Tissue from 8 animals (4 males and 4 females) for each biological replicate of the prestin line and 4 animals (2 males and 2 females) for each biological replicate of the Sox2 line were processed for ribosome immunoprecipitation (5 μg of purified anti-HA.11, BioLegend) followed by RNA extraction using the RNeasy Plus Micro kit (QIAGEN) as previously described (Song et al., 2018). RNA quality was determined using an Agilent Technologies Bioanalyzer 2100 RNA pico chip as per the manufacturer’s instructions (Agilent Technologies). All samples had a RIN score above 8.
Reverse transcription and real-time PCR
Efficiency of the ribosome immunoprecipitation was assessed by reverse transcription followed by real time PCR. Equal amounts of RNA from the input and the RiboTag-IP samples were used for reverse transcription using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific). The real time PCR was performed on an Applied Biosystems® StepOnePlus Real-Time PCR System with the Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific) and the following primers: Gapdh-Fw 5′-GGAGAAACCTGCCAAGTATGA-3′; Gapdh-Rv 5′- TCCTCAGTGTAGCCCAAGA-3′; Sox2-Fw 5′-CCCACCTACAGCATGTCCTA-3′; Sox2-Rv 5′-GTGGGAGGAAGAGGTAACCA-3′; Slc26a5-Fw 5′-GAAAGGCCCATCTTCAGTCATC-3′; Slc26a5-Rv 5′-GCCACTTAGTGATAGGCAGGAAC-3′; Pou3f4-Fw 5′-CTGCCTCGAATCCCTACAGC-3′; Pou3f4-Rv 5′-CTGCAAGTAGTCACTTTGGAGAA-3′.
tdTomato expression
Mice were euthanized and their temporal bones removed, fixed with 4% paraformaldehyde (PFA) (Alfa Aesar) overnight at 4°C and incubated in EDTA 0.5M until adequate decalcification. Cochlear ducts were dissected, permeabilized with PBS-0.3% Triton X-100 before addition of Alexa Fluor 488® phalloidin (1:1000, Thermo Fisher Scientific) to stain F-actin. Tissue was mounted on slides with ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Images were acquired using a Zeiss 5Live & Zeiss 510 at the University of Maryland School of Medicine Center for Innovative Biomedical Resources Confocal Microscopy facility – Baltimore, Maryland.
Cytocochleogram
Following the final ABR recording, mice were euthanized, and their temporal bones removed and fixed with 4% PFA (Alfa Aesar) overnight at 4°C. The temporal bones were then immersed into 0.5M EDTA at 4°C until adequate decalcification. Following decalcification, the cochlear ducts were dissected to expose the Organ of Corti. Dissected tissues were permeabilized for 1 hour in PBS-0.5% TWEEN 20 and blocked for 1 hour in permeabilization buffer supplemented with 5% normal donkey serum (Sigma-Aldrich) at room temperature. Tissues were then incubated with a goat polyclonal anti-prestin (1:200, Santa-Cruz Biotechnology) at 4°C overnight, followed by incubation with a donkey anti-goat IgG Alexa Fluor® 546 (1:800, Thermo Fisher Scientific). The nuclei were counterstained with 300nM of DAPI and the tissue was mounted with the ProLong Gold antifade reagent (Thermo Fisher Scientific). Fluorescence images of the outer hair cell were captured using a Nikon Eclipse E600 microscope (20x objective) equipped with an Infinity 3 camera (Lumenera). Following image capture, cochlear frequencies were mapped using the Measure Line ImageJ plugin developed by the Eaton-Peabody Laboratories at the Massachusetts Eye and Ear (available for download at https://rsbweb.nih.gov/ij/). Outer hair cells were counted along the basilar membrane from the apex toward the base between frequency intervals of 1 to 2 kHz. Hair cell counts were performed up to 53 kHz (the equivalent of 85% from the apex [Müller et al., 2005]) as up to this frequency intact samples could be collected from all biological replicates.
RNA quantification using nanoString technology
RNA extraction was performed as described for the ribosome IP, from one additional replicate for each condition. RNA was processed for nanoString at the UMSOM Institute for Genome Sciences using the nCounter Standard Master Kit (nanoString) following the manufacturer’s instructions. Quantification was performed using the nanoString nCounter platform and data were analyzed using the nSolver 4.0 software. The probe IDs from nanoString are listed in Table S2.
Flow cytometry for CD45+ immune cells
To prepare single cell suspension, cochleae from n = 6 mice per experimental condition were pooled and digested in 1xAccutase (Stem Cell Technology) for 20 min in 37°C, washed with staining buffer containing 1% fetal bovine serum (FBS) in PBS, centrifuged at 400 g for 5 min at 4°C and plated in a 96-well plate. The cells were blocked with CD16/CD32 (1:200, Thermo Fisher Scientific) and stained with live dead cell marker using Aqua Live/Dead stain kit (1:1000, Amcyan) in the presence of RNase inhibitor (1:400, Promega) in PBS for 15 min. After subsequent wash, the cells obtained from each condition were stained with a 1:1 mixture of CD45-PE (1:200, 30F11, Biolegend) and a corresponding TotalSeq antibody (Biolegend) in staining buffer containing RNase inhibitor (1:400) for 30 min. The following antibodies were used for each condition: TotalSeq™ A0301-Hashtag1 antibody (M1/42, 30F11) for Sham exposed cells, TotalSeq™ A0302-Hashtag2 antibody (M1/42, 30F11) for 3d, TotalSeq™ A0303-Hashtag3 antibody (M1/42, 30F11) for 7d and TotalSeq™ A0304-Hashtag4 antibody (M1/42, 30F11) for 14d post noise. CD45 positive living cells were sorted on BDFACS ARIA III (BD Biosciences) in 500 μL collecting buffer containing RNase free 1% BSA (Thermo Fischer Scientific) and RNase inhibitor (1:400). The cells were centrifuged at 400 g for 5 min at 4°C and resuspended in 50 μL collecting buffer.
Cell dissociation of spiral ganglion neurons
Dissection and dissociation of SGN-biased samples from naive (n = 4) and noise exposed (n = 4) 9-week-old-male CBA/CaJ was done as previously described (Sun et al., 2018). Briefly, temporal bones mice were isolated, and the overlying bone and lateral wall were extracted leaving just the modiolus, with the spiral ganglion, the spiral limbus, inner sulcus, and portions of the organ of Corti and outer sulcus remaining. Microdissected tissue from each mouse was pooled and processed for dissociation using collagenase IV (Thermo Fisher Scientific) and DNase I (Stem Cell Technologies), followed by incubation with papain (Worthington Biochemical) and mechanical disruption by trituration. After addition of 20% ovomucoid protease inhibitor (Worthington Biochemical), the cell suspension was passed through a 20 μm filter (pluriSelect) to remove debris. The cells were washed twice with PBS containing 0.04% BSA (PBS/BSA) and resuspended in PBS/BSA solution. An aliquot was used to count the cells on a Luna FL automated counter using an AO/PI fluorescent cell viability assay (Logos Biosystems). Time from euthanasia to single-cell capture was ~3h.
Cell dissociation of the lateral wall
Temporal bones from naive (n = 4) and noise exposed (n = 4) 7–8 wko, male CBA/CaJ mice were isolated in ice-cold Leibovitz’s L-15 medium. Using a #11 scalpel, the bony wall of the cochlea was removed to reveal the cochlear lateral wall. The lateral wall with attached stria vascularis was extracted from apex to base and separated from the modiolus and other cochlear tissue. Microdissected tissue was then dissociated into single cells using the collagenase IV and papain enzyme mixtures as described for the dissociation of SGN tissue. For the lateral wall tissues, however, the total incubation time in papain was extended to 1 hour to ensure complete tissue dissociation.
In situ hybridization using RNAscope
Tissue for RNAscope was obtained from male and female B6CBAF1/J mice (F1 progeny of a cross between C57BL/6J females and CBA/J males, Jackson Laboratory, Stock No: 100011). Mice were non-exposed or exposed to noise at 10 weeks of age (n = 2 males and 1 female for each condition) as described in the “Noise Exposure.” Animals were euthanized by CO2 asphyxiation at 24h post noise and the inner ears collected. Tissue was incubated in RNase free 4% paraformaldehyde (Alfa Aesar) at 4°C overnight, followed by decalcification in 150 mM EDTA (Quality Biological) at 4°C for 3 days. Decalcified inner ears were incubated in a sucrose gradient, embedded in Super Cryoembedding Medium (SCEM) (Section-Lab), flash frozen in liquid nitrogen and kept at −80°C until sectioning. Twelve μm sections were obtained using a CM 1850 cryostat (Leica). In situ hybridization was performed using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics) following the manufacturer’s instructions with the probes listed in the key resource table. Images of 0.2 μm sections to create a Z stack were taken using a Nikon W1 spinning disk confocal on Nikon Ti2 inverted microscope (40x oil objective) at the University of Maryland School of Medicine Center for Innovative Biomedical Resources Confocal Microscopy Facility – Baltimore, Maryland. Images were then processed using the Fiji (ImageJ) program (Schindelin et al., 2012). Quantification of the RNA dots was performed using QuPath version 0.2.3 (Bankhead et al., 2017). The number of neurons was obtained using the ‘Cell detection’ tool followed by the ‘Subcellular detection’ tools to quantify individual and clustered dots corresponding to RNA molecules.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-HA.11 | BioLegend | Cat# 901513; RRID:AB_2565335 |
| anti-prestin | Santa-Cruz Biotechnology | Cat# sc-22692; RRID:AB_2302038 |
| donkey anti-goat IgG Alexa Fluor® 546 | Thermo Fisher Scientific | Cat# A-11056; RRID:AB_2534103 |
| CD45-PE (30F11) | BioLegend | Cat# 103106; RRID:AB_312971 |
| TotalSeq™ A0301-Hashtag1 antibody (M1/42; 30-F11) | BioLegend | Cat# 155801; RRID:AB_2750032 |
| TotalSeq™ A0302-Hashtag2 antibody (M1/42; 30-F11) | BioLegend | Cat# 155803; RRID:AB_2750033 |
| TotalSeq™ A0303-Hashtag3 (M1/42; 30-F11) | BioLegend | Cat# 155805; RRID:AB_2750034 |
| TotalSeq™ A0304-Hashtag4 (M1/42; 30-F11) | BioLegend | Cat# 155807; RRID:AB_2750035 |
| CD16/CD32 Monoclonal Antibody | Thermo Fisher Scientific | Cat# 14–0161-82; RRID:AB_467133 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| KetaVed (Ketamine) | Covetrus | Cat# 048858 |
| Keatminol (Ketamine) | Intervet | Cat# 511485 |
| AnaSed (Xylazine) | Covetrus | Cat# 033197 |
| Rompun (Xylazine) | Bayer | Cat# KP0A43D |
| Alexa Fluor 488® phalloidin | Thermo Fisher Scientific | Cat# A12379 |
| AO/PI stain | Logos Biosystems | Cat# F23001 |
| DynaBeads Dynabeads MyOne Silane | Thermo Fisher Scientific | Cat# 37002D |
| SPRI beads | Beckman Coulter | Cat# B23318 |
| Super Cryoembedding Medium (SCEM) | Section-Lab | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| NEBNext® Ultra Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7420 |
| nCounter Standard Master Kit | NanoString Technologies | Cat# NAA-AKIT-048 |
| Chromium single cell 3′reagent kit V3 | 10x Genomics | Cat# PN-1000075 |
| Single Cell 3′ Reagent Kits v2 | 10x Genomics | Cat# PN-120237 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostics | Cat# 323100 |
|
| ||
| Deposited data | ||
|
| ||
| RiboTag raw datasets | This paper | GEO: GSE168973 |
| SGN and LW raw datasets | This paper | GEO: GSE168041 |
| CD45+ raw dataset | This paper | GEO: GSE167078 |
| Analyzed datasets | This paper | https://umgear.org/p?l=76ae3595 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Ai14 | The Jackson Laboratory | Stock No: 007914; RRID:IMSR_JAX:007914 |
| Mouse: RiboTag | Kindly provided by Dr. MK Lobo, University of Maryland Baltimore | RRID:IMSR_JAX:011029 |
| Mouse: Prestin-CreERT2 | Kindly provided by Dr J Zuo St. Jude Children’s Research Hospital | RRID:MGI:5316666 |
| Mouse: Sox2-CreERT2 | The Jackson Laboratory | Stock No: 017593; RRID:IMSR_JAX:017593 |
| Mouse: CBA/CaJ | The Jackson Laboratory | Stock No: 000654; RRID:IMSR_JAX:000654 |
| Mouse: B6CBAF1/J | The Jackson Laboratory | Stock No: 100011; RRID:IMSR_JAX:100011 |
| Mouse: CBA/Ca/Sca | Scanbur—*N.B. colony has been terminated by distributor | Cat# CBSSIMA0 |
| Mouse: Sox2CreERT2/+;RiboTagHA/+; CBA/CaJ | This paper | N/A |
| Mouse: PrestinCreERT2/+;RiboTagHA/+; CBA/CaJ | This paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| RNAscope® Probe- Mm-Atf3 | Advanced Cell Diagnostics | Cat# 426891 |
| RNAscope® Probe- Mm-Atf4 | Advanced Cell Diagnostics | Cat# 405101 |
| RNAscope® Probe- Mm-Ddit3-C3 | Advanced Cell Diagnostics | Cat# 317661-C3 |
| RNAscope® Probe- Mm-Gadd45a-C2 | Advanced Cell Diagnostics | Cat# 460571-C2 |
| RNAscope® Probe- Mm-Runx1-C3 | Advanced Cell Diagnostics | Cat# 406671-C3 |
| RNAscope® Probe- Mm-Vgf | Advanced Cell Diagnostics | Cat No. 517421 |
| Oligonucleotides, standard desalting | Integrated DNA Technologies | Sequences can be found in detailed methods |
| Oligonucleotides designed by nanoString | Integrated DNA Technologies | Sequence IDs can be found in Table S2 |
|
| ||
| Software and algorithms | ||
|
| ||
| BioSigRP | Tucker-Davis Technologies | RRID:SCR_014590 |
| nSolver 4.0 software | NanoString Technologies | RRID:SCR_003420 |
| Cell Ranger | 10x Genomics | RRID:SCR_017344 |
| DESeq2 | Bioconductor | RRID:SCR_015687 |
| DAVID | https://david.abcc.ncifcrf.gov/ | RRID:SCR_001881 |
| Seurat v3 | https://satijalab.org/seurat/ | RRID:SCR_016341 |
| MAST | Bioconductor | RRID:SCR_016340 |
| biomaRt | Bioconductor | RRID:SCR_019214 |
| Fiji | (Schindelin et al., 2012) | RRID:SCR_002285 |
| QuPath | Bankhead et al., 2017 | RRID:SCR_018257 |
| gene Expression Analysis Resource (gEAR) | https://umgear.org/ | RRID:SCR_017467 |
| BioRender | https://biorender.com/ | RRID:SCR_018361 |
|
| ||
| Other | ||
|
| ||
| RZ6 Processor | Tucker-Davis Technologies | N/A |
| RX6 Processor | Tucker-Davis Technologies | N/A |
| 1/4” microphone | PCB Piezotronics | Model PCB-378C01 |
| 1/4” microphone | Brüel and Kjær | Models 4939 A 011 and 2690 A 0S1 |
| Microphone | Knowles | Model EK 23103 |
| Acoustic coupler with a pre-amplifier | Etymotic Research | Model ER-10B+ |
| Chromium Controller | 10x Genomics | N/A |
| GemCode Platform | 10x Genomics | N/A |
Library preparations and sequencing
RiboTag samples
The RNA samples were processed for library preparation using the NEBNext® Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs). The libraries were sequenced on an HiSeq 4000 Sequencing System (Illumina) with a configuration of 75 base read length and pair-end reads.
CD45+ immune cell samples
To generate gene expression libraries from single cells, Chromium single cell 3′reagent kit v3 (10x Genomics) was used according to the manufacturer’s instructions. The number of cells were not counted as the cell numbers were low, but a quality check with Tryptan blue was performed on the remnant cells in the tube after the cell suspension was loaded onto the chip. Briefly, cells were encapsulated into droplets, lysed, and reverse transcribed with barcodes (53°C for 45 min; 85°C for 5 min) in a 96-well Thermal cycler (Thermo Fisher Scientific). After breaking the droplets, barcoded cDNA was purified with Dynabeads MyOne Silane (Thermo Fisher Scientific) and PCR-amplified in 11 cycles (98°C for 3 min; [98°C for 15, 63°C for 20 s, 72°C for 1 min] x 11 cycles; 72°C for 1 min. Since the cells were hash tagged, the Hash Tag Oligonucleotides (Integrated DNA Technologies; custom DNA oligos) were added to the cDNA Amplification Reaction Mix at volume of 1 μL of a 0.2 μM primer mix. Subsequently, the amplified cDNA was fragmented, ligated with adapters, sample-indexed and selected with SPRI beads (Beckman Coulter) to average 400 bp size for gene expression library and 180 bp for HTO library. The resulting cDNA library was sequenced on Illumina NovaSeq 6000 platform.
SGN and lateral wall samples
Single cell capture, library preparation, and sequencing were performed as previously described (Sun et al., 2018). A target capture of 10,000 cells per sample was chosen using the high throughput, droplet microfluidics GemCode platform from 10x Genomics with v2 chemistry (Zheng et al., 2017). Each droplet contains a cell and a gel bead hybridized with oligo(dT) primers encoding a unique cell barcode and unique molecular identifiers (UMIs) in lysis buffer. Following capture, the transcriptomes captured on gel beads are pooled and reverse transcribed to cDNA. Reverse transcription and PCR amplification of cDNA, as well as the preparation of a library from 3′ ends were conducted according to the manufacturer’s published protocol. We performed 15 cycles of PCR amplification of cDNA. The library was sequenced on an Illumina NovaSeq 6000 at the Broad Institute Sequencing Facility. Reads were demultiplexed, aligned to the GRCm38 mm10 assembly reference genome, and filtered; and cell barcodes and UMIs were quantified using the Cell Ranger pipeline with default parameters (https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome). Cell Ranger uses STAR (Dobin et al., 2013) for alignment and manufacturer’s software for all other steps (Zheng et al., 2017).
Bioinformatic analyses
RiboTag datasets
To focus the analysis on genes that were robustly detected, only those which were covered by at least 40 reads in all replicates of any condition were considered in our analysis. Counts were than transformed to rpkm units, which normalize the reads by transcript length allowing for cross comparison between genes, and then further normalized using quantile normalization. Enrichment factor (EF) in the RiboTag-IP samples compared to the input samples was calculated as log2 of the ratio between the corresponding average levels. To avoid inflation of EF estimates (due to low level in input samples) we set a floor level that was equal to the 10th percentile of the expression level distribution. All levels below this floor level were set to this level. We found that EFs were systematically correlated with transcript length. We used Lowess normalization to correct for this technical effect (Mandelboum et al., 2019). We defined the set of OHC-enriched genes as those showing (1) OHC-EF greater than 1.5 (that is, genes showing > 2.83 ( = 21.5) enrichment in the OHC RiboTag-IP compared to the input sample) and (2) greater than 2-fold enrichment in OHCs compared to SCs (that is, ΔEF = OHC-EF – SC-EF > 1). 436 genes met these criteria. Using analogous criteria, 248 genes were called as SC-enriched genes. Differentially expressed (DE) genes were detected using DESeq2 (Love et al., 2014). In addition to FDR < 5% we required that DE genes show clear separation in expression level between the two conditions. Specifically, if a gene showed elevated expression in condition.A compared to condition.B, we required that the lowest level measured in condition.A is at least 1.2-fold higher than the highest level measure for that gene in condition.B. This added criterion significantly increases the confidence of the called DEGs at the cost of decreased sensitivity. We elected to add this criterion to increase the stringency of the results. Four out of 36 samples (from Sox2-CreERT2;RiboTag;CBA/CaJ: 1 input and IP baseline samples, 1 input and 1 IP 6h samples) were excluded because of sequencing technical issues. One Prestin-CreERT2;Ribotag;CBA/CaJ baseline IP sample was detected as an outlier and was excluded from further analyses. Merging resulting OHC and SC datasets we used DESeq2′s regularized log transformation (rlog) on the original count data, followed by quantile normalization. Cluster analysis was applied to the union of the DEGs detected in either the OHC or SC dataset. It was done using the CLuster Identification via Connectivity Kernels (CLICK) algorithm (with default parameters) implemented in the EXpression Analyzer and DisplayER (EXPANDER) package (Hait et al., 2019). In the combined analysis of the OHC and SC datasets, two additional Prestin-CreERT2;RiboTag;CBA/CaJ samples (1 input and 1 IP at 6h) were excluded as they appeared as outlier in the principal component analysis. Clustering was done on fold-change levels (in log2), were calculated in each condition relative to its baseline condition. To focus on response pattern (rather than on magnitude), fold-change levels of each gene (row) were standardized to mean = 0 and SD = 1. For the integrated cluster analysis of the OHC and SC datasets, to increased homogeneity of the reported clusters, we filtered each cluster to retain only genes whose pattern was highly correlated (r > 0.8) with the cluster’s mean pattern. Figures 1J and 1K shows the filtered clusters. GO enrichment analysis was done using Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2009a, 2009b).
SGN scRNA-seq dataset
Using Seurat v3 (Butler et al., 2018), count matrices from all samples were merged into one matrix of 34,776 cells and 22,292 genes. Dissociation of cochlea cells often results in red blood cells from outside the cochlea contaminating the sample. As these cells highly express hemoglobin genes (Hba-a1, Hba-a2, Hbb-bh1, Hbb-bs, Hbb-bt), we filtered out the 546 cells (0.02% of the cells) with 1% or more reads from hemoglobin genes. In addition, we filtered out cells with more than 10% reads from mitochondrial genes and outlier cells with more than 7,000 or less than 500 detected genes (Figure S2A), remaining with 25,994 cells. We kept the 17,662 genes that were expressed in at least 20 cells (median genes detected per cell - 2,165). Clustering was carried using sctransform pipeline implement by Seurat (Hafemeister and Satija, 2019). Briefly, for normalization of gene counts, we used the Pearson residuals from a regularized negative binomial regression adding the sequencing depth of the cells as a covariate. The dimensional reduction of the expression matrix was achieved by first selecting the 3,000 (sctransform default) most highly variable genes (HVG) (most variable Person residuals). The HVG residuals-cells matrix was transformed using principal component analysis (PCA) and the top 25 principal components were kept. For clustering, Euclidean distances between cells in this dimensionally reduced space were used to construct a shared nearest neighbor graph. This graph was used to cluster the cells using the modularity optimization algorithm implemented by Seurat. Uniform Manifold Approximation and Projection (UMAP) was used to visualize the dimensionally reduced space. For analysis of marker genes, e.g., tables, violin plot and UMAP visualizations of expression, we renormalized the gene counts using Seurat default normalization (log normalization). This normalization divides gene counts for each cell by the total counts for that cell and multiplies by 10,000. This is then transformed using a natural log (adding a pseudo count of 1). We identified clusters representing neuronal and Schwann cells according to known marker genes (Figure S2B). To achieve clustering resolution that allows proper separation between neuron subtypes in the SGN, we isolated the 9,327 cells assigned to these clusters and repeated dimensional reduction and clustering analysis. The UMAP indicated that cells were clustered largely according to the experimental condition (noise or control) (Figure S2C). Therefore, to enable clustering that better reflects cell identity, we merged cells from the noise-exposed and control samples using an anchor-based integration pipeline implemented by Seurat (Stuart et al., 2019). Briefly, Canonical correlation analysis (CCA) was used to transform a set of 2,000 (default) genes that are most variable in both conditions. A graph constructed based on the top 30 CCA components was used to identify and rank anchors – pairs of control-noise cells that are mutual nearest neighbors. Then the expression of the most variable genes was adjusted using a transformation designed to minimize variability between pairs in anchors. The adjusted expression levels obtained from integration were used for dimensional reduction, clustering, and UMAP visualization as described above. Following this integration procedure, we were able to identify type 1, type 2 and Schwann clusters, but we also identified one cluster, which expressed SGN genes (Nefh, Nefl, Tubb3), but was separated from the other SGN clusters, and showed high expression of Coch, a gene known to be highly expressed in fibrocytes (Robertson et al., 2001) (Figures S2D and S2E). We removed the 411 cells of this cluster and repeated the analysis once more. The UMAP confirmed that cells did not cluster according to treatment (Figure S2F) or individual mice (Figure S2G). As above, for inspection of marker gene expression, we used log normalization (and not the transformed expression values used for data integration). The marker genes showed similar expression levels in cells from the control and noise-exposed cells demonstrating that these cell types were confidently defined in both conditions (Figures S2H–S2J). For analysis of immune cells, we revisited the clustering of 25,994 cells that passed quality criteria. We isolated 1,499 cells from the clusters specifically expressing monocytes/macrophages and neutrophiles marker genes (Figure S5J). The anchor-based approach described above was used to re-cluster these cells (Figure 5D; Figure S5K).
Lateral wall scRNA-seq dataset
We merged the expression data from all eight samples and obtained a matrix with 36,933 cells and 21,470 genes. We kept the 34,341 cells with less than 1% reads from hemoglobin genes (99.9% of the cells), less than 25% reads from mitochondrial genes, and with 1,000 – 6,000 detected genes (median genes detected per cell - 2,872) (Figure S4A). We filtered out genes expressed in less than 20 cells and left with 16,832 genes. Normalization, dimensional reduction, clustering, and data integration were carried as described above for the SGN dataset. Cell clusters corresponding to basal, intermediate, spindle/root cells, B cells, fibrocyte, monocyte, and neutrophils were identified based on the expression of known marker genes (Figures S4B and S4C). We then isolated the 26,259 cells of these eight clusters and repeated dimensional reduction and clustering. Final clusters are shown in Figure 4A. For analysis of immune cells, we revisited the clustering of 34,341 cells that passed quality criteria. We isolated 655 cells from the clusters specifically expressing monocytes/macrophages and neutrophils marker genes (Figure S5N). The anchor-based approach described above was used to re-cluster these cells (Figure 5E; Figure S5P). Inspecting the results of this analysis showed a cluster of 53 cells with outlier percentage of reads from mitochondrial genes (average of 9.5% in comparison to 4.1% for the rest of the cells) (Figure S5O). We removed these cells and repeated the clustering analysis.
CD45+ cells scRNA-seq dataset
Cells were sequenced following a procedure that utilizes cell hashing with barcoded antibodies for multiplexing (Stoeckius et al., 2018). We used an antibody directed against CD45 to conjugated Hashtag oligonucleotides (HTOs), assigning a distinct HTO to each of the four conditions (baseline, 3, 7 and 14 days). Subsequently, the transcriptomic RNA and HTOs from cells of all conditions were separately sequenced. The Cell Ranger Software Suite (Version 3.0.2) was used for de-multiplexing the samples, to perform barcode processing and single cell 3′UMI counting using mouse mm10 as reference genome. Thus, Cell ranger output consisted of a 31,053 by 1,556 genes-cells matrix and a 4 by 1,556 HTO-cells matrix. We used Seurat utilities for analysis of Cell Hashing datasets to assign condition (HTO) for each cell and to identify cell barcodes corresponding to droplets with zero (negatives) or multiple cells (doublets). As expected, inferred negatives, singlets, and doublets displayed different distributions of UMI counts (Figure S5A); singlets and doublets appeared well separated in tSNE visualization based on gene expression (Figure S5B); and cells specifically expressed their assigned HTO (Figure S5C), indicating a proper assignment of HTOs to cells. We filtered out the 152 cells that were classified as negatives and the 152 that were classified as doublets. Out of the singlets, we kept 1,123 cells with less than 7% reads from mitochondrial genes, and with more than 500 and less than 6,000 detected genes (Figure S5D). Normalization, dimensional reduction, and clustering were carried as described above for the other datasets. Cell clusters did not show any gross impact for condition/HTO identity of the cells (Figure S5E). We were not able to differentiate between monocytes and macrophages based on marker genes, and therefore refer to these immune cells as monocytes or monocytes/macrophages.
Differential expression for scRNA-seq datasets
Differential expression analysis was done using Model-based Analysis of Single Cell Transcriptomics (MAST) (Finak et al., 2015). It was applied to each cell cluster separately, comparing the noise-exposed and control cells. The number of detected genes in each cell was added as a covariate to MAST model. Only genes expressed in at least 10% of the cells in either condition were tested. A gene was considered differentially expressed if its FDR q-value was less than 0.05 and its absolute fold change was greater than 1.2. Hierarchical clustering of the DEGs were done using Ward’s method implemented by R hclust function. As hierarchical clustering cannot be performed with too many missing values (which in our case, resulted from instances where a gene was not sufficiently expressed in a cell type to obtain a fold change value from MAST analysis), we replaced missing values with a value of 0.
GO-terms enrichment, hearing loss genes, motif and cell communication analyses for scRNA-seq datasets
For GO-term enrichment analysis, the hypergeometric test implemented by the enricher function from clusterProfile R package was used (Yu et al., 2012). Gene sets were obtained from msigdbr R package. For each cell type, the tested target set consisted of either the up- or downregulated DEGs, while the background set consisted of all the genes that were tested by MAST for that cell type. GO terms with FDR q-value < 0.05 were considered enriched (A cutoff of q-value < 0.1 was used for the analysis of Srebf1 targets [Figure S3G]). For the gene set enrichment analysis of hearing loss genes, we used the function GSEA from clusterProfile package. For each cell type, we rank all its genes according to their log fold change (noise versus control), so that upregulated genes are at the top of the list and downregulated at the bottom. The input to GSEA function was, for each cell type, the ranked list of genes, and one of the hearing loss genes lists as the gene set. RcisTarget R package (Aibar et al., 2017) was used for motif analysis, using its gene-motif rankings for mouse genes which is based on sequences 500 bp upstream the TSSs. For analysis of cell-cell signaling, we used CellPhoneDB (Efremova et al., 2020) with default parameters. As CellPhoneDB is based on human genes, we converted the mouse genes to their human ortholog, using biomaRt (Durinck et al., 2009) and keeping only genes with one-to-one Hs-Mm ortholog mapping. Potential communication between cells is detected by CellPhone based on the expression of a receptor by one cell type and its ligand by another cell type. To identify candidate drugs that could modify the inner ear response to noise, we intersected the drug-target interaction data from DrugCentral (Ursu et al., 2017) with the list of the DEGs identified in all cell types. Keeping interactions of FDA-approved drugs and of high target activity level (TCRD target development levels classified as ‘Tchem’ or ‘Tclin’), we identified 2,936 DEG/cell-drug pairs from 897 drugs. We next sorted the list of DEG/cell-drug pairs by the number of genes targeted per drug. Finally, we curated this list by counting only drugs in which their effect opposes the gene expression kinetics measured in response to PTS-inducing noise, in at least 50% of the interactions (Table S7).
QUANTIFICATION AND STATISTICAL ANALYSIS
Comparison of ABR thresholds before and after noise exposure within a group were analyzed by a two-way ANOVA or a Mixed-effects analysis with Tukey’s post hoc test for multiple comparisons using Prism 9 software (GraphPad, CA) for the different time points. Comparisons of OHC loss before and after noise were performed using Student’s t test assuming unequal variance using Microsoft Excel. Statistical details for each experiment can be found in the figure legends. Significance was defined as p < 0.05 and q < 0.05.
Supplementary Material
Highlights.
A cell-type-specific transcriptomic map of the cochlear response to noise
Noise-resilient type 1A auditory neurons upregulate the ATF3/4 pathway
Monocytes significantly alter their gene expression in response to noise exposure
STAT3/IRF7 are probable regulators of a general cochlear transcriptomic response to noise
ACKNOWLEDGMENTS
The authors would like to thank Hela Azaiez, PhD, and Mr. Mark Wieber for technical support (R.H.), SciLife for assisting with the scRNA-seq (B.C.), Ms. Evangelia Tserga for assisting with the collection of tissues for the scRNA-seq (B.C.), and the Eukaryotic Single Cell Genomics facility at Science for Life Laboratory in Stockholm (B.C.). This work was funded by NIDCD/NIH R01DC013817 and R01DC03544, DOD CDMRP MR130240 and RH200052, Carolyn Frenkil Foundation, and the Hearing Restoration Project (HRP) of the Hearing Health Foundation (R.H.); the Swedish Medical Research Council and Hörselforskningsfonden (B.C.); the Karolinska Institutet, Tysta Skolan, and Office of the Assistant Secretary of Defense for Health Affairs, through the Neurosensory and Rehabilitation under award no. W81XWH-16–1-0032 (B.C. and C.R.C.); and the European Union’s Horizon 2020 research and innovation programme nos. 722046 and 848261 (C.R.C). This work was also supported by The United States - Israel Binational Science Foundation (2017218; R.H. and R.E.); The Edmond J. Safra Center for Bioinformatics at Tel Aviv University (E.D.S. and R.E.); and Teva Pharmaceutical Industries, Ltd. and the Israeli National Forum for BioInnovators (E.D.S.). Some illustrations in the graphical abstract were created with BioRender (https://biorender.com/).
Footnotes
DECLARATION OF INTERESTS
K.S.S., J.B.S., G.P., A.T.P., and J.B. are employees of Decibel Therapeutics. The data presented in this manuscript are registered for pending U.S. Provisional Patent Application, number: 63/151,249, title: “System and Methods for Cell Type-Specific Atlas for the Inner Ear Transcriptional Response to Acoustic Trauma,” and UMB docket number: RH-2021-073.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109758.
REFERENCES
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amici SA, Dunn WA Jr., Murphy AJ, Adams NC, Gale NW, Valenzuela DM, Yancopoulos GD, and Notterpek L (2006). Peripheral myelin protein 22 is in complex with α6β4 integrin, and its absence alters the Schwann cell basal lamina. J. Neurosci. 26, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, and Hochedlinger K (2011). Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Yoo JE, Choe YH, Kwak SH, Choi JY, Jung J, and Hyun Y-M (2021). Neutrophils infiltrate into the spiral ligament but not the stria vascularis in the cochlea during lipopolysaccharide-induced inflammation. Theranostics 11, 2522–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, et al. (2018). Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS ONE 13, e0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. (2017). QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen A, Anderson L, Bakay W, and Forge A (2019). Localized disorganization of the cochlear inner hair cell synaptic region after noise exposure. Biol. Open 8, bio038547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Collado MS, Oliver ER, and Corwin JT (2013). Specializations of intercellular junctions are associated with the presence and absence of hair cell regeneration in ears from six vertebrate classes. J. Comp. Neurol. 521, 1430–1448. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Vethanayagam RR, Yang S, Bard J, Jamison J, Cartwright D, Dong Y, and Hu BH (2014). Molecular profile of cochlear immunity in the resident cells of the organ of Corti. J. Neuroinflammation 11, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederroth CR, Canlon B, and Langguth B (2013). Hearing loss and tinnitus–are funders and industry listening? Nat. Biotechnol. 31, 972–974. [DOI] [PubMed] [Google Scholar]
- Cederroth CR, Park JS, Basinou V, Weger BD, Tserga E, Sarlus H, Magnusson AK, Kadri N, Gachon F, and Canlon B (2019). Circadian Regulation of Cochlear Sensitivity to Noise by Circulating Glucocorticoids. Curr. Biol. 29, 2477–2487.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Peppi M, Kujawa SG, and Sewell WF (2009). Regulated expression of surface AMPA receptors reduces excitotoxicity in auditory neurons. J. Neurophysiol. 102, 1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Chung CH, Lu CH, and Chien WC (2019). Metformin decreases the risk of sudden sensorineural hearing loss in patients with diabetes mellitus: A 14-year follow-up study. Diab. Vasc. Dis. Res. 16, 324–327. [DOI] [PubMed] [Google Scholar]
- Chessum L, Matern MS, Kelly MC, Johnson SL, Ogawa Y, Milon B, McMurray M, Driver EC, Parker A, Song Y, et al. (2018). Helios is a key transcriptional regulator of outer hair cell maturation. Nature 563, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JW, Ahn JH, Kim JY, Lee HJ, Kang HH, Lee YK, Kim JU, and Koo SW (2007). The effect of isoflurane, halothane and pentobarbital on noise-induced hearing loss in mice. Anesth. Analg. 104, 1404–1408. [DOI] [PubMed] [Google Scholar]
- Clough E, and Barrett T (2016). The Gene Expression Omnibus database. Methods in Molecular Biology (Humana Press), pp. 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate TM, Scott MK, and Gurjar M (2019). Current concepts in cochlear ribbon synapse formation. Synapse 73, e22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M, Agerman K, Ernfors P, and Canlon B (2000). Complementary roles of neurotrophin 3 and a N-methyl-D-aspartate antagonist in the protection of noise and aminoglycoside-induced ototoxicity. Proc. Natl. Acad. Sci. USA 97, 7597–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M, Vento-Tormo M, Teichmann SA, and Vento-Tormo R (2020). CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506. [DOI] [PubMed] [Google Scholar]
- Fang J, Zhang W-C, Yamashita T, Gao J, Zhu M-S, and Zuo J (2012). Outer hair cell-specific prestin-CreERT2 knockin mouse lines. Genesis 50, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez KA, Jeffers PWC, Lall K, Liberman MC, and Kujawa SG (2015). Aging after noise exposure: acceleration of cochlear synaptopathy in “recovered” ears. J. Neurosci. 35, 7509–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez KA, Watabe T, Tong M, Meng X, Tani K, Kujawa SG, and Edge ASB (2021a). Trk agonist drugs rescue noise-induced hidden hearing loss. JCI Insight 6, e142572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez KA, Allen P, Campbell M, Page B, Townes T, Li CM, Cheng H, Garrett J, Mulquin M, Clements A, et al. (2021b). Atorvastatin is associated with reduced cisplatin-induced hearing loss. J. Clin. Invest. 131, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye MD, Ryan AF, and Kurabi A (2019). Inflammation associated with noise-induced hearing loss. J. Acoust. Soc. Am. 146, 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman AC, Kujawa SG, and Liberman MC (2013). Noise-induced cochlear neuropathy is selective for fibers with low spontaneous rates. J. Neurophysiol. 110, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2019 Hearing Loss Collaborators (2021). Hearing loss prevalence and years lived with disability, 1990–2019: Findings from the Global Burden of Disease Study 2019. Lancet. 397, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafidi A (1998). Peripherin-like immunoreactivity in type II spiral ganglion cell body and projections. Brain Res. 805, 181–190. [DOI] [PubMed] [Google Scholar]
- Hait TA, Maron-Katz A, Sagir D, Amar D, Ulitsky I, Linhart C, Tanay A, Sharan R, Shiloh Y, Elkon R, and Shamir R (2019). The EXPANDER Integrated Platform for Transcriptome Analysis. J. Mol. Biol. 431, 2398–2406. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Hickman TT, Suzuki J, Ji L, Kohrman DC, Corfas G, and Liberman MC (2019). Protection from noise-induced cochlear synaptopathy by virally mediated overexpression of NT3. Sci. Rep. 9, 15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranen A, Ikäheimo K, Virkkala J, and Pirvola U (2018). The Stress Response in the Non-sensory Cells of the Cochlea Under Pathological Conditions-Possible Role in Mediating Noise Vulnerability. J. Assoc. Res. Otolaryngol. 19, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R, Lipford EL, and Depireux D (2020a). Noise: Acoustic Trauma to the Inner Ear. Otolaryngol. Clin. North Am. 53, 531–542. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Gwilliam K, Rose KP, Milon B, and Matern MS (2020b). Cell Type-Specific Expression Analysis of the Inner Ear: A Technical Report. Laryngoscope, Published online June 24, 2020. 10.1002/lary.28765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman TT, Hashimoto K, Liberman LD, and Liberman MC (2020). Synaptic migration and reorganization after noise exposure suggests regeneration in a mature mammalian cochlea. Sci. Rep. 10, 19945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, and Liberman MC (2003). Lateral wall histopathology and endocochlear potential in the noise-damaged mouse cochlea. J. Assoc. Res. Otolaryngol. 4, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa M, Olszewski R, Li X, Taukulis I, Gu S, DeTorres A, Lopez IA, Linthicum FH Jr., Ishiyama A, Martin D, et al. (2020). Characterizing Adult Cochlear Supporting Cell Transcriptional Diversity Using Single-Cell RNA-Seq: Validation in the Adult Mouse and Translational Implications for the Adult Human Cochlea. Front. Mol. Neurosci. 13, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Rutherford MA, and Green SH (2020). Protection of cochlear synapses from noise-induced excitotoxic trauma by blockade of Ca2+-permeable AMPA receptors. Proc. Natl. Acad. Sci. USA 117, 3828–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hunt D, Raivich G, and Anderson PN (2012). Activating transcription factor 3 and the nervous system. Front. Mol. Neurosci. 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongkamonwiwat N, Ramirez MA, Edassery S, Wong ACY, Yu J, Abbott T, Pak K, Ryan AF, and Savas JN (2020). Noise Exposures Causing Hearing Loss Generate Proteotoxic Stress and Activate the Proteostasis Network. Cell Rep. 33, 108431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesici GG, Öcal FCA, Gürgen SG, Erdem ŞR, Öğüş E, Erbek HS, and Özlüoğlu LN (2018). The protective effect of metformin against the noise-induced hearing loss. Eur. Arch. Otorhinolaryngol. 275, 2957–2966. [DOI] [PubMed] [Google Scholar]
- Kim JU, Lee HJ, Kang HH, Shin JW, Ku SW, Ahn JH, Kim YJ, and Chung JW (2005). Protective effect of isoflurane anesthesia on noise-induced hearing loss in mice. Laryngoscope 115, 1996–1999. [DOI] [PubMed] [Google Scholar]
- Kim KX, Payne S, Yang-Hood A, Li SZ, Davis B, Carlquist J, V-Ghaffari B, Gantz JA, Kallogjeri D, Fitzpatrick JAJ, et al. (2019). Vesicular glutamatergic transmission in noise-induced loss and repair of cochlear ribbon synapses. J. Neurosci. 39, 4434–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleshchevnikov V, Shmatko A, Dann E, Aivazidis A, King HW, Li T, Lomakin A, Kedlian V, Jain MS, Park JS, et al. (2020). Comprehensive mapping of tissue cell architecture via integrated single cell and spatial transcriptomics. bioRxiv. 10.1101/2020.11.15.378125. [DOI] [Google Scholar]
- Kolla L, Kelly MC, Mann ZF, Anaya-Rocha A, Ellis K, Lemons A, Palermo AT, So KS, Mays JC, Orvis J, et al. (2020). Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 11, 2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korrapati S, Taukulis I, Olszewski R, Pyle M, Gu S, Singh R, Griffiths C, Martin D, Boger E, Morell RJ, and Hoa M (2019). Single Cell and Single Nucleus RNA-Seq Reveal Cellular Heterogeneity and Homeostatic Regulatory Networks in Adult Mouse Stria Vascularis. Front. Mol. Neurosci. 12, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzing MK, Luo V, and Firestein BL (2012). Protection from glutamate-induced excitotoxicity by memantine. Ann. Biomed. Eng. 40, 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, and Kiang NY (1978). Acoustic trauma in cats. Cochlear pathology and auditory-nerve activity. Acta Otolaryngol. Suppl. 358, 1–63. [PubMed] [Google Scholar]
- Liberman MC, and Kujawa SG (2017). Cochlear synaptopathy in acquired sensorineural hearing loss: Manifestations and mechanisms. Hear. Res. 349, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LD, Suzuki J, and Liberman MC (2015). Dynamics of cochlear synaptopathy after acoustic overexposure. J. Assoc. Res. Otolaryngol. 16, 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Kariya S, Uraguchi K, Takahara J, Fujimoto S, Sugaya A, and Nishizaki K (2021). Immediate changes in transcription factors and synaptic transmission in the cochlea following acoustic trauma: A gene transcriptome study. Neurosci. Res. 165, 6–13. [DOI] [PubMed] [Google Scholar]
- Mandelboum S, Manber Z, Elroy-Stein O, and Elkon R (2019). Recurrent functional misinterpretation of RNA-seq data caused by sample-specific gene length bias. PLoS Biol. 17, e3000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson DR (2018). Sensory hair cells: An introduction to structure and physiology. Integr. Comp. Biol. 58, 282–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon B, Mitra S, Song Y, Margulies Z, Casserly R, Drake V, Mong JA, Depireux DA, and Hertzano R (2018). The impact of biological sex on the response to noise and otoprotective therapies against acoustic injury in mice. Biol. Sex Differ. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzack EL, and Cunningham LL (2013). Lead roles for supporting actors: critical functions of inner ear supporting cells. Hear. Res. 303, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, von Hünerbein K, Hoidis S, and Smolders JWT (2005). A physiological place-frequency map of the cochlea in the CBA/J mouse. Hear. Res. 202, 63–73. [DOI] [PubMed] [Google Scholar]
- Oishi N, Kendall A, and Schacht J (2014). Metformin protects against gentamicin-induced hair cell death in vitro but not ototoxicity in vivo. Neurosci. Lett. 583, 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis J, Gottfried B, Kancherla J, Adkins RS, Song Y, Dror AA, Olley D, Rose K, Chrysostomou E, Kelly MC, et al. (2021). gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat. Methods 18, 843–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, and Gorman AM (2016). The integrated stress response. EMBO Rep. 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Kim SW, Park K, Choung YH, Jou I, and Park SM (2012). Pravastatin attenuates noise-induced cochlear injury in mice. Neuroscience 208, 123–132. [DOI] [PubMed] [Google Scholar]
- Peixoto Pinheiro B, Vona B, Löwenheim H, Rüttiger L, Knipper M, and Adel Y (2021). Age-related hearing loss pertaining to potassium ion channels in the cochlea and auditory pathway. Pflugers Arch. 473, 823–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins RE, and Morest DK (1975). A study of cochlear innervation patterns in cats and rats with the Golgi method and Nomarkski Optics. J. Comp. Neurol. 163, 129–158. [DOI] [PubMed] [Google Scholar]
- Petitpré C, Wu H, Sharma A, Tokarska A, Fontanet P, Wang Y, Helmbacher F, Yackle K, Silberberg G, Hadjab S, and Lallemend F (2018). Neuronal heterogeneity and stereotyped connectivity in the auditory afferent system. Nat. Commun. 9, 3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel JL, d’Aldin C, Ruel J, Ladrech S, and Pujol R (1997). Synaptic repair mechanisms responsible for functional recovery in various cochlear pathologies. Acta Otolaryngol 117, 214–218. [DOI] [PubMed] [Google Scholar]
- Puel JL, Ruel J, Gervais d’Aldin C, and Pujol R (1998). Excitotoxicity and repair of cochlear synapses after noise-trauma induced hearing loss. Neuroreport 9, 2109–2114. [DOI] [PubMed] [Google Scholar]
- Rai V, Wood MB, Feng H, Schabla NM, Tu S, and Zuo J (2020). The immune response after noise damage in the cochlea is characterized by a heterogeneous mix of adaptive and innate immune cells. Sci. Rep. 10, 15167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson NG, Resendes BL, Lin JS, Lee C, Aster JC, Adams JC, and Morton CC (2001). Inner ear localization of mRNA and protein products of COCH, mutated in the sensorineural deafness and vestibular disorder, DFNA9. Hum. Mol. Genet. 10, 2493–2500. [DOI] [PubMed] [Google Scholar]
- Ruel J, Wang J, Pujol R, Hameg A, Dib M, and Puel JL (2005). Neuroprotective effect of riluzole in acute noise-induced hearing loss. Neuroreport 16, 1087–1090. [DOI] [PubMed] [Google Scholar]
- Ruel J, Wang J, Rebillard G, Eybalin M, Lloyd R, Pujol R, and Puel JL (2007). Physiology, pharmacology and plasticity at the inner hair cell synaptic complex. Hear. Res. 227, 19–27. [DOI] [PubMed] [Google Scholar]
- Ryan AF, Kujawa SG, Hammill T, Le Prell C, and Kil J (2016). Temporary and Permanent Noise-induced Threshold Shifts: A Review of Basic and Clinical Observations. Otol. Neurotol. 37, e271–e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryugo DK, and Parks TN (2003). Primary innervation of the avian and mammalian cochlear nucleus. Brain Res. Bull. 60, 435–456. [DOI] [PubMed] [Google Scholar]
- Sadler E, Ryals MM, May LA, Martin D, Welsh N, Boger ET, Morell RJ, Hertzano R, and Cunningham LL (2020). Cell-Specific Transcriptional Responses to Heat Shock in the Mouse Utricle Epithelium. Front. Cell. Neurosci. 14, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, and Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA 106, 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilder AGM, Su MP, Blackshaw H, Lustig L, Staecker H, Lenarz T, Safieddine S, Gomes-Santos CS, Holme R, and Warnecke A (2019). Hearing Protection, Restoration, and Regeneration: An Overview of Emerging Therapeutics for Inner Ear and Central Hearing Disorders. Otol. Neurotol. 40, 559–570. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seist R, Tong M, Landegger LD, Vasilijic S, Hyakusoku H, Katsumi S, McKenna CE, Edge ASB, and Stankovic KM (2020). Regeneration of Cochlear Synapses by Systemic Administration of a Bisphosphonate. Front. Mol. Neurosci. 13, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Liu L, He T, Guo X, Yu Z, Yin S, and Wang J (2013). Ribbon synapse plasticity in the cochleae of Guinea pigs after noise-induced silent damage. PLoS ONE 8, e81566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Liu K, Wang H, Zhang Y, Hong Z, Wang M, Wang X, Jiang X, and Yang S (2015). Noise induced reversible changes of cochlear ribbon synapses contribute to temporary hearing loss in mice. Acta Otolaryngol. 135, 1093–1102. [DOI] [PubMed] [Google Scholar]
- Shi L, Chang Y, Li X, Aiken SJ, Liu L, and Wang J (2016). Coding deficits in noise-induced hidden hearing loss may stem from incomplete repair of ribbon synapses in the cochlea. Front. Neurosci. 10, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha BR, Chia C, Wu L, Kujawa SG, Liberman MC, and Goodrich LV (2018). Sensory Neuron Diversity in the Inner Ear Is Shaped by Activity. Cell 174, 1229–1246.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Milon B, Ott S, Zhao X, Sadzewicz L, Shetty A, Boger ET, Tallon LJ, Morell RJ, Mahurkar A, and Hertzano R (2018). A comparative analysis of library prep approaches for sequencing low input translatome samples. BMC Genomics 19, 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM 3rd, Smibert P, and Satija R (2018). Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Liu J, Crary JF, Malagelada C, Sulzer D, Greene LA, and Levy OA (2013). ATF4 protects against neuronal death in cellular Parkinson’s disease models by maintaining levels of parkin. J. Neurosci. 33, 2398–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Babola T, Pregernig G, So KS, Nguyen M, Su SM, Palermo AT, Bergles DE, Burns JC, and Müller U (2018). Hair Cell Mechanotransduction Regulates Spontaneous Activity and Spiral Ganglion Subtype Specification in the Auditory System. Cell 174, 1247–1263.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghibiglou C, Martin HGS, Lai TW, Cho T, Prasad S, Kojic L, Lu J, Liu Y, Lo E, Zhang S, et al. (2009). Role of NMDA receptor-dependent activation of SREBP1 in excitotoxic and ischemic neuronal injuries. Nat. Med. 15, 1399–1406. [DOI] [PubMed] [Google Scholar]
- Ursu O, Holmes J, Knockel J, Bologa CG, Yang JJ, Mathias SL, Nelson SJ, and Oprea TI (2017). DrugCentral: online drug compendium. Nucleic Acids Res. 45 (D1), D932–D939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, Robin C, and van Oudenaarden A (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936. [DOI] [PubMed] [Google Scholar]
- Walters BJ, Yamashita T, and Zuo J (2015). Sox2-CreER mice are useful for fate mapping of mature, but not neonatal, cochlear supporting cells in hair cell regeneration studies. Sci. Rep. 5, 11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Corfas G, and Stone JS (2013). Inner ear supporting cells: rethinking the silent majority. Semin. Cell Dev. Biol. 24, 448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangemann P (2002). K+ cycling and the endocochlear potential. Hear. Res. 165, 1–9. [DOI] [PubMed] [Google Scholar]
- Warchol ME (2019). Interactions between macrophages and the sensory cells of the inner ear. Cold Spring Harb. Perspect. Med. 9, a033555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T, Omelchenko I, Foster S, Zhang Y, Shi X, and Nuttall AL (2014). JAK2/STAT3 inhibition attenuates noise-induced hearing loss. PLoS ONE 9, e108276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, O’Malley JT, de Gruttola V, and Liberman MC (2020). Age-related hearing loss is dominated by damage to inner ear sensory cells, not the cellular battery that powers them. J. Neurosci. 40, 6357–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Shen W, He DZZ, Long KB, Madison LD, and Dallos P (2000). Prestin is the motor protein of cochlear outer hair cells. Nature 405, 149–155. [DOI] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data generated during this study have been deposited in the Gene Expression Omnibus and are available under accession numbers GEO: GSE168041 (SGN and lateral wall scRNA-seq), GEO: GSE167078 (CD45+ scRNA-seq) and GEO: GSE168973 (Prestin-CreERT2;RiboTag and Sox2-CreERT2;RiboTag). All of the analyzed data are also available for browsing and analysis via the gene Expression Analysis Resource (umgear.org; https://umgear.org/NIHL).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.