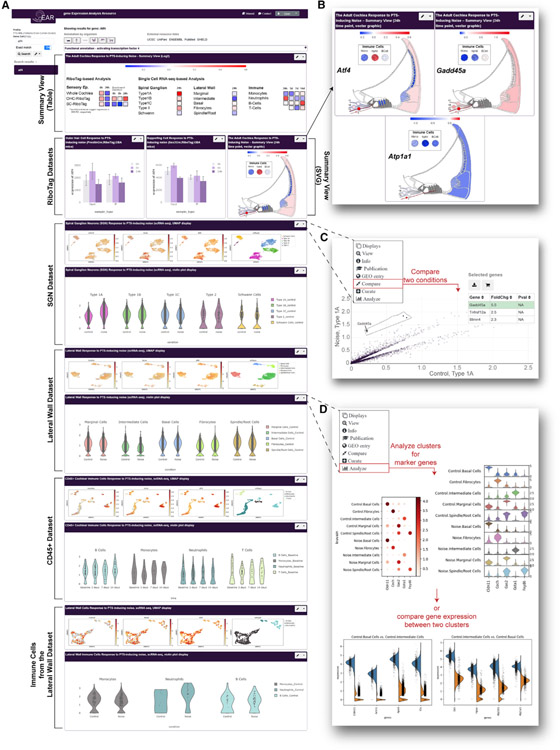
Figure 7. Data sharing, visualization, and analysis via the gEAR (umgear.org).
A custom profile was generated in the gEAR to support sharing, visualization, and analysis of the processed transcriptomic data presented as part of this manuscript (https://umgear.org/NIHL).
(A) Overview of the manuscript profile, which contains two summary views (tabular and graphic, depicting gene expression as log2 fold change), the OHC and SC RiboTag datasets (bar graphs), and the SGN, LW, and immune cells datasets (UMAPs and violin plots).
(B) Examples of the graphic summary view representation for Atf4, Gadd45a, and Atp1a1 showing the (log2) fold change in gene expression following noise exposure mapped onto anatomical sites for intuitive interpretation of the data.
(C and D) The gEAR portal contains several analysis tools allowing users to further explore the data in the cloud.
(C) Example of the “compare tool,” which enables users to compare expression across any two conditions within a single dataset, here showing the DEGs in type 1A SGNs between control and noise samples.
(D) The single-cell workbench allows users to perform “de novo” analysis of the data or use a “stored analysis” to explore marker genes and compare across clusters. Here are shown marker genes for the 5 clusters (before and after noise) of the LW dataset (top) and the top 4 DEGs between intermediate and basal cells (bottom).