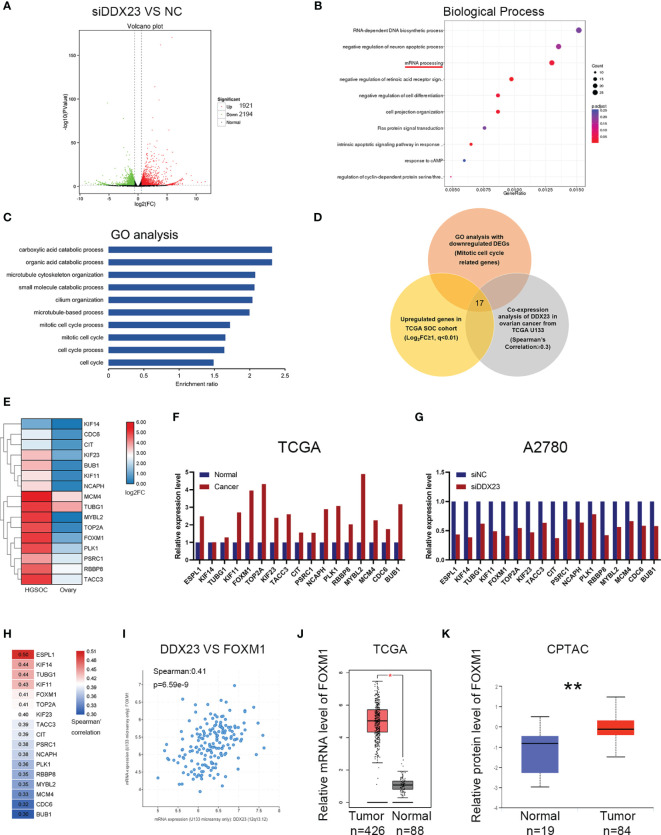
Figure 6.
Identification of differentially expressed genes involved in DDX23 function by RNA-seq. (A) Volcano plot for the DEGs between siDDX23 and NC groups (1.5-FC, P < 0.05). (B) Biological process analysis showed that DDX23 was involved in mRNA processing. (C) GO enrichment analysis of the downregulated DEGs between siDDX23 and NC groups. (D) Venn diagram of 17 hub genes obtained by overlapping three gene sets. Gene set 1, 123 downregulated genes related to mitotic cell cycle processes identified by RNA-seq; Gene set 2, upregulated genes in ovarian cancer from GEPIA (Log2FC ≥ 1, q < 0.01); Gene set 3, genes that positively related to DDX23 expression (TCGA U133 microarray, Spearman’s Correlation ≥ 0.3). (E, F) Relative mRNA expression of 17 candidate downstream genes in TCGA database. (G) qRT-PCR analysis of mRNA expression of 17 candidates in A2780 cells. (H) Co-expression analysis between DDX23 and 17candidates expression in ovarian cancer based on the cBioPortal database. (I) Co-expression analysis between DDX23 and FOXM1 expression in ovarian cancer based on the cBioPortal database. (J) FOXM1 mRNA expression in ovarian cancer and normal ovary samples in TCGA cohort from GEPIA. (K) FOXM1 protein expression in ovarian cancer and normal ovary samples in CPTAC cohort. DEG, differential expression gene; NC, negative control; FC, fold change; GO, Gene Ontology; GEPIA, Gene Expression Profiling Interactive Analysis; TCGA, The Cancer Genome Atlas. *P < 0.05, **P < 0.01.