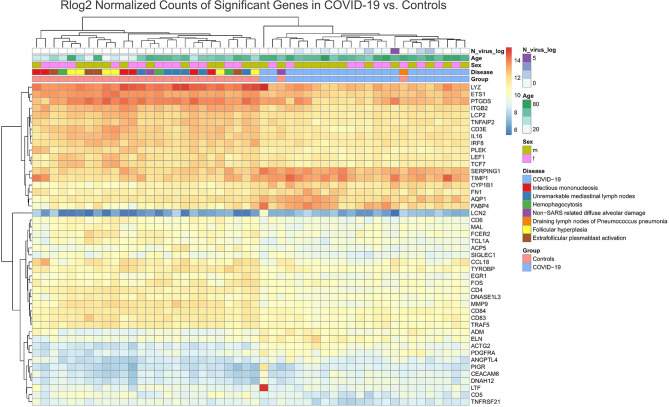
Figure 7.
Unsupervised Cluster Analysis of Differentially Expressed Genes in the Draining Lymph Nodes of COVID-19 Compared to Control Cases. A downregulation of genes related to T-cell function and differentiation (LCP2, CD3E, LEF1, CD6, MAL, TCL1A, TYROBP, CD84, CD5), in particular CD4 associated (IL16, CD4) and genes encoding for immunoglobulin receptors (FCER2, PIGR) and transcription factors (ETS1, EGR1, FOS) was observed in COVID-19 patients versus controls. A downregulation of LYZ and SIGLEC1 indicates an altered macrophage phenotype (less prevalent M1 polarization). Genes related to complement and fluid homeostasis and vascular functions (SERPING1, AQP1, ADM, ANGPTL4) are upregulated. A dysregulation in expression amongst genes encoding for leukocyte adhesion and motility was observed (downregulation of IRGB2, upregulation of ACTG2 and DNASH12). MMP9 is downregulated while its inhibitor TIMP1 is upregulated. Other genes involved in remodeling (ELN, PDGFRA and FN1) are upregulated. A downregulation of PTGDS and upregulation of CYP1B1 indicates an altered arachidonic acid metabolism.