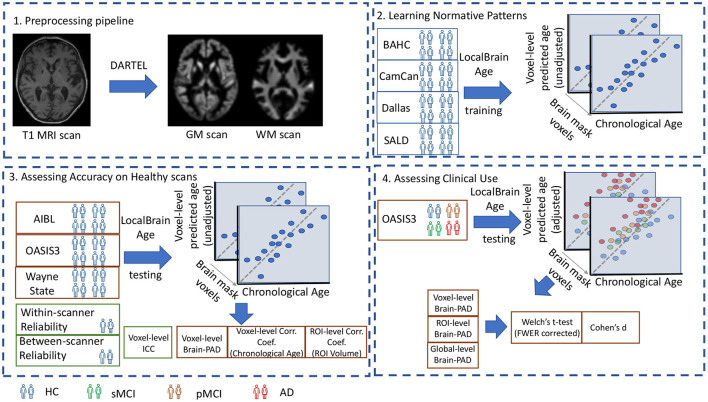
Figure 1.
1. Preprocessing pipeline: T1-weighted MRI scans from all datasets were tissue segmented and non-linearly registered using SPM12 to generate modulated grey and white matter volume maps. 2. Learning Normative Patterns: Randomly subsampled participants from BAHC, CamCan, Dallas, and SALD were used to train the local brain-age U-net to learn predict chronological age locally. 3. Assessing Accuracy on Healthy scans: Using healthy scans from AIBL, OASIS3 and Wayne State we tested age prediction accuracy on unseen data from independent datasets. 4. Assessing Clinical Use: Using the OASIS3 dataset (healthy controls, stable MCI, progressive MCI, and AD participants), we compared groups at voxel, regional and global levels.