Figure 4.
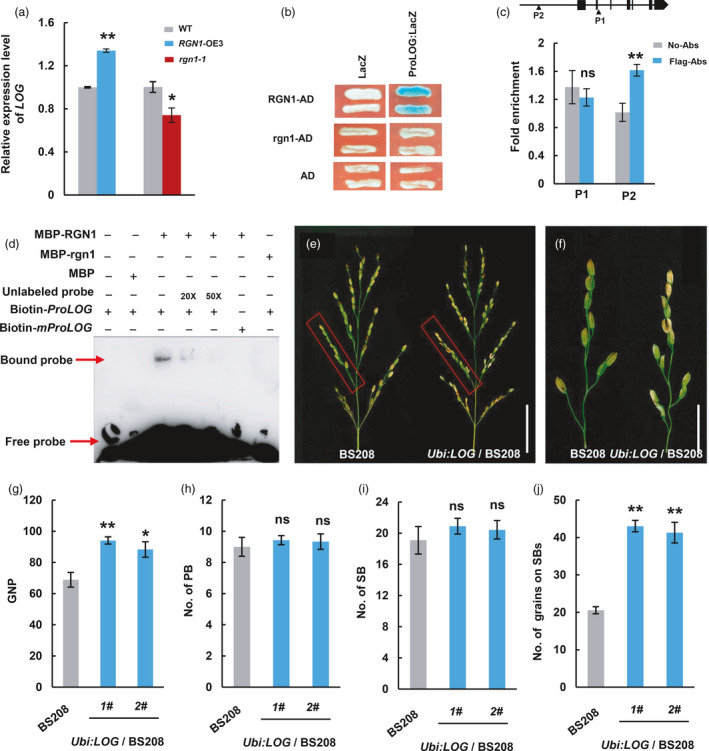
RGN1 controls LOG expression to regulate the formation of lateral grains on secondary branches. (a) Expression levels of LOG in NIP compared to rgn1‐1 and RGN1‐OE3 plants. Values are means ± SEM, (n = 3, each with three technical repeats). (b) Yeast one‐hybrid assays demonstrate that RGN1 binds to the LOG promoter (ProLOG). (c) Quantitative ChIP‐PCR indicates that RGN1 binds to the MS188‐binding motif (P2 region) from the LOG promoter in vivo. Values are means ± SEM, (n = 3). (d) Electrophoretic mobility shift assay shows that RGN1 binds to the MS188‐binding motif from the LOG promoter in vitro. mProLOG, LOG promoter containing mutated MS188 binding motif. (e,f) Phenotype of BS208 was partially rescued by LOG overexpression: panicles architecture (e), primary branches (f) from (e) within red boxes of BS208 and Ubi:LOG/BS208 transgenic plant. Scale bars, 5 cm for (e); 2 cm for (f). (g–j) Comparisons of grain number per panicle (g), primary branch number (h), secondary branches number (i), number of grains on secondary branches (j) between BS208 and Ubi:LOG/BS208 transgenic plants. Values are means ± SEM (n = 10). *P < 0.05, **P < 0.01, ns, no significant difference. Two‐tailed student’s t‐tests.
